Biología molecular del cáncer colorrectal
Molecular biology of colorectal cancer
Cómo citar
Descargar cita

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Mostrar biografía de los autores
Introducción: el cáncer colorrectal es una de las principales causas de incidencia y mortalidad por cáncer a nivel mundial. Su desarrollo resulta de una compleja interacción entre alteraciones genéticas, epigenéticas y del microambiente tumoral.
Métodos: se realizó una revisión narrativa de la literatura científica relevante sobre la biología molecular del cáncer colorrectal, incluyendo vías de carcinogénesis, síndromes hereditarios, biomarcadores clínicamente relevantes y aplicaciones de la biopsia líquida.
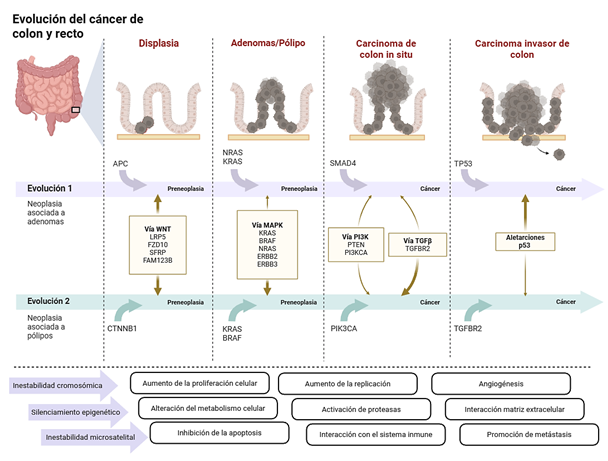
Resultados: la patogénesis del cáncer colorrectal se organiza en tres vías principales de inestabilidad genómica: inestabilidad cromosómica, caracterizada por alteraciones en APC, TP53 y activación de RAS/MAPK; inestabilidad de microsatélites, secundaria a deficiencia del sistema de reparación de errores de emparejamiento, asociada a alta carga mutacional y sensibilidad a inmunoterapia; y el fenotipo metilador de islas CpG, frecuentemente relacionado con mutaciones en BRAF y silenciamiento de MLH1. Aproximadamente el 10% de los casos corresponde a síndromes hereditarios, principalmente síndrome de Lynch y síndromes polipósicos. Biomarcadores moleculares permiten estratificación pronóstica y selección terapéutica, mientras que el ADN tumoral circulante emerge como herramienta para detección de enfermedad mínima residual y monitorización de la evolución clonal.
Conclusión: la integración de la biología molecular en el manejo del cáncer colorrectal constituye un pilar de la oncología de precisión, optimizando el diagnóstico, el pronóstico y la selección de terapias personalizadas.
Visitas del artículo 0 | Visitas PDF 0
Descargas
- Bray F, Ferlay J, Laversanne M, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin [Internet]. 2024;130(3):474–498. Disponible en: https://doi.org/10.3322/caac.21834
- Morgan E, Arnold M, Gini A, et al. Global burden of colorectal cancer in 2020 and 2040: incidence and mortality estimates from GLOBOCAN. Gut [Internet]. 2023;72(2):338–344. Disponible en: https://doi.org/10.1136/gutjnl-2022-327736
- Nguyen HT, Duong HQ. The molecular characteristics of colorectal cancer: implications for diagnosis and therapy. Oncol Lett [Internet]. 2018;16(1):9–18. Disponible en: https://doi.org/10.3892/ol.2018.8679
- Li Q, Geng S, Luo H, et al. Signaling pathways involved in colorectal cancer: pathogenesis and targeted therapy. Signal Transduct Target Ther [Internet]. 2024;9(1):266. Disponible en: https://doi.org/10.1038/s41392-024-01953-7
- Cisyk AL, Nugent Z, Wightman RH, et al. Characterizing microsatellite instability and chromosome instability in interval colorectal cancers. Neoplasia [Internet]. 2018;20(9):943–950. Disponible en: https://doi.org/10.1016/j.neo.2018.07.007
- Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature [Internet]. 2012;487(7407):330–337. Disponible en: https://doi.org/10.1038/nature11252
- Stoffel EM, Boland CR. Genetics and genetic testing in hereditary colorectal cancer. Gastroenterology [Internet]. 2015;149(5):1191–1203.e2. Disponible en: https://doi.org/10.1053/j.gastro.2015.07.021
- Punt CJ, Koopman M, Vermeulen L. From tumour heterogeneity to advances in precision treatment of colorectal cancer. Nat Rev Clin Oncol [Internet]. 2017;14(4):235–246. Disponible en: https://doi.org/10.1038/nrclinonc.2016.171
- Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA by ultradeep sequencing in patients with stages I to III colorectal cancer. JAMA Oncol [Internet]. 2019;5(8):1124–1131. Disponible en: https://doi.org/10.1001/jamaoncol.2019.0528
- Fearon ER, Vogelstein B. A genetic model for colorectal tumorigenesis. Cell [Internet]. 1990;61(5):759–767. Disponible en: https://doi.org/10.1016/0092-8674(90)90186-I
- Markowitz SD, Bertagnolli MM. Molecular origins of cancer: molecular basis of colorectal cancer. N Engl J Med[Internet]. 2009;361(25):2449–2460. Disponible en: https://doi.org/10.1056/NEJMra0804588
- Al-Sohaily S, Biankin A, Leong R, et al. Molecular pathways in colorectal cancer. J Gastroenterol Hepatol[Internet]. 2012;27(9):1423–1431. Disponible en: https://doi.org/10.1111/j.1440-1746.2012.07200.x
- Clevers H, Nusse R. Wnt/β-catenin signaling and disease. Cell [Internet]. 2012;149(6):1192–1205. Disponible en: https://doi.org/10.1016/j.cell.2012.05.012
- Koveitypour Z, Panahi F, Vakilian M, et al. Signaling pathways involved in colorectal cancer progression. Cell Biosci [Internet]. 2019;9:97. Disponible en: https://doi.org/10.1186/s13578-019-0361-4
- Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol [Internet]. 2011;42(1):1–10. Disponible en: https://doi.org/10.1016/j.humpath.2010.06.002
- Guinney J, Dienstmann R, Wang X, et al. The consensus molecular subtypes of colorectal cancer. Nat Med[Internet]. 2015;21(11):1350–1356. Disponible en: https://doi.org/10.1038/nm.3967
- Seshagiri S, Stawiski EW, Durinck S, et al. Recurrent R-spondin fusions in colon cancer. Nature [Internet]. 2012;488(7413):660–664. Disponible en: https://doi.org/10.1038/nature11282
- Glaviano A, Foo ASC, Lam HY, et al. PI3K/AKT/mTOR signaling transduction pathway and targeted therapies in cancer. Mol Cancer [Internet]. 2023;22(1):138. Disponible en: https://doi.org/10.1186/s12943-023-01827-6
- Appleyard JW, Williams CJM, Manca P, et al. Targeting the MAP kinase pathway in colorectal cancer: a journey in personalized medicine. Clin Cancer Res [Internet]. 2025;31(13):2565–2572. Disponible en: https://doi.org/10.1158/1078-0432.CCR-25-0107
- Massagué J. TGF-β in cancer. Cell [Internet]. 2008;134(2):215–230. Disponible en: https://doi.org/10.1016/j.cell.2008.07.001
- Grady WM, Markowitz SD. Genetic and epigenetic alterations in colon cancer. Annu Rev Genomics Hum Genet[Internet]. 2002;3:101–128. Disponible en: https://doi.org/10.1146/annurev.genom.3.022502.103043
- Puzzo M, De Santo M, Morelli C, et al. Colorectal cancer: current and future therapeutic approaches and related technologies addressing multidrug strategies against multiple-level resistance mechanisms. Int J Mol Sci [Internet]. 2025;26(3):1313. Disponible en: https://doi.org/10.3390/ijms26031313
- Chen Y, Zheng X, Wu C. The role of the tumor microenvironment and treatment strategies in colorectal cancer. Front Immunol [Internet]. 2021;12:792691. Disponible en: https://doi.org/10.3389/fimmu.2021.792691
- Wu X, Yan H, Qiu M, et al. Comprehensive characterization of tumor microenvironment in colorectal cancer via molecular analysis. eLife [Internet]. 2023;12:e86032. Disponible en: https://doi.org/10.7554/eLife.86032
- Terzić J, Grivennikov S, Karin E, et al. Inflammation and colon cancer. Gastroenterology [Internet]. 2010;138(6):2101–2114.e5. Disponible en: https://doi.org/10.1053/j.gastro.2010.01.058
- Veenendaal LM, Kranenburg O, Smakman N, et al. Differential Notch and TGF-β signaling in primary colorectal tumors and their corresponding metastases. Cell Oncol [Internet]. 2008;30(1):1–11. Disponible en: https://doi.org/10.1155/2008/839076
- Eng C, Yoshino T, Ruíz-García E, et al. Colorectal cancer. Lancet [Internet]. 2024;404(10449):294–310. Disponible en: https://doi.org/10.1016/S0140-6736(24)00360-X
- Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: a review. JAMA [Internet]. 2021;325(7):669–685. Disponible en: https://doi.org/10.1001/jama.2021.0106
- Goosenberg E, Kaur A, Babiker HM. A review of hereditary colorectal cancers. En: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing; 2025. Disponible en: https://www.ncbi.nlm.nih.gov/books/NBK538195/
- Stjepanovic N, Moreira L, Carneiro F, et al. Hereditary gastrointestinal cancers: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol [Internet]. 2019;30(10):1558–1571. Disponible en: https://doi.org/10.1093/annonc/mdz233
- Lynch HT, de la Chapelle A. Hereditary colorectal cancer. N Engl J Med [Internet]. 2003;348(10):919–932. Disponible en: https://doi.org/10.1056/NEJMra012242
- Abdel-Rahman WM, Peltomäki P. Molecular basis and diagnostics of hereditary colorectal cancers. Ann Med [Internet]. 2004;36(5):379–388. Disponible en: https://doi.org/10.1080/07853890410018222
- Curtius K, Gupta S, Boland CR. Lynch syndrome: a mechanistic and clinical management update. Aliment Pharmacol Ther [Internet]. 2022;55(8):960–977. Disponible en: https://doi.org/10.1111/apt.16826
- Taieb J, Svrcek M, Cohen R, Basile D, Tougeron D, Phelip JM. Deficient mismatch repair/microsatellite unstable colorectal cancer: diagnosis, prognosis and treatment. Eur J Cancer [Internet]. 2022;175:136–157. Disponible en: https://doi.org/10.1016/j.ejca.2022.07.020
- Colas C, Guerrini-Rousseau L, Suerink M, et al. ERN GENTURIS guidelines on constitutional mismatch repair deficiency: diagnosis, genetic counselling, surveillance, quality of life, and clinical management. Eur J Hum Genet[Internet]. 2024;32(12):1526–1541. Disponible en: https://doi.org/10.1038/s41431-024-01708-6
- Mur P, García-Mulero S, Del Valle J, et al. Role of POLE and POLD1 in familial cancer. Genet Med [Internet]. 2020;22(12):2089–2100. Disponible en: https://doi.org/10.1038/s41436-020-0922-2
- Palles C, Martin L, Domingo E, et al. The clinical features of polymerase proofreading-associated polyposis and recommendations for patient management. Fam Cancer [Internet]. 2022;21(2):197–209. Disponible en: https://doi.org/10.1007/s10689-021-00256-y
- Ditonno I, Novielli D, Celiberto F, et al. Molecular pathways of carcinogenesis in familial adenomatous polyposis. Int J Mol Sci [Internet]. 2023;24(6):5687. Disponible en: https://doi.org/10.3390/ijms24065687
- Joo JE, Viana-Errasti J, Buchanan DD, et al. Genetics, genomics and clinical features of adenomatous polyposis. Fam Cancer [Internet]. 2025;24(2):38. Disponible en: https://doi.org/10.1007/s10689-025-00460-0
- Zare B, Monahan KJ. Guidelines for familial adenomatous polyposis: challenges in defining clinical management for a rare disease. Fam Cancer [Internet]. 2025;24(2):35. Disponible en: https://doi.org/10.1007/s10689-025-00462-y
- Zaffaroni G, Mannucci A, Koskenvuo L, et al. Updated European guidelines for clinical management of familial adenomatous polyposis (FAP), MUTYH-associated polyposis (MAP), gastric adenocarcinoma, proximal polyposis of the stomach (GAPPS) and other rare adenomatous polyposis syndromes: a joint EHTG-ESCP revision. Br J Surg [Internet]. 2024;111(5):znae070. Disponible en: https://doi.org/10.1093/bjs/znae263
- Luo P, Shi W, Cheng X, et al. Which drugs are more effective in preventing familial adenomatous polyposis progression based on network meta-analysis? Curr Pharm Des [Internet]. 2024;30(20):1548–1563. Disponible en: https://doi.org/10.2174/0113816128289465240422074745
- Farooq U, El Alayli A, Duvvuri A, et al. Nonsteroidal anti-inflammatory drugs for chemoprevention in patients with familial adenomatous polyposis: a systematic review and meta-analysis. Gastro Hep Adv [Internet]. 2023;2(7):1005–1013. Disponible en: https://doi.org/10.1016/j.gastha.2023.05.009
- Dunlop MG, Farrington SM. MUTYH-associated polyposis and colorectal cancer. Surg Oncol Clin N Am [Internet]. 2009;18(4):599–610. Disponible en: https://doi.org/10.1016/j.soc.2009.08.003
- Nielsen M, Infante E, Brand R. MUTYH Polyposis. En: Adam MP, Feldman J, Mirzaa GM, Pagon RA, Wallace SE, Amemiya A, et al., editores. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2025. Actualizado 2021 May 27. Disponible en: https://www.ncbi.nlm.nih.gov/books/NBK107219
- Yehia L, Plitt G, Tushar AM, et al. Extended spectrum of cancers in PTEN hamartoma tumor syndrome. NPJ Precis Oncol [Internet]. 2025;9(1):61. Disponible en: https://doi.org/10.1038/s41698-025-00847-3
- Yehia L, Eng C. PTEN Hamartoma Tumor Syndrome. En: Adam MP, Mirzaa GM, Pagon RA, Wallace SE, Bean LJH, Gripp KW, et al., editores. GeneReviews® [Internet]. Seattle (WA): University of Washington, Seattle; 1993–2021. Disponible en: https://www.ncbi.nlm.nih.gov/books/NBK1488/
- Vasen HFA, Tomlinson I, Castells A. Clinical management of hereditary colorectal cancer syndromes. Nat Rev Gastroenterol Hepatol [Internet]. 2015;12(2):88–97. Disponible en: https://doi.org/10.1038/nrgastro.2014.229
- Jaeger E, Leedham S, Lewis A, et al. Hereditary mixed polyposis syndrome is caused by a 40-kb upstream duplication that leads to increased and ectopic expression of the BMP antagonist GREM1. Nat Genet [Internet]. 2012;44(6):699–703. Disponible en: https://doi.org/10.1038/ng.2263
- Gala MK, Mizukami Y, Le LP, Moriichi K, et al. Germline mutations in oncogene-induced senescence pathways are associated with multiple sessile serrated adenomas. Gastroenterology [Internet]. 2014;146(2):520–529. Disponible en: https://doi.org/10.1053/j.gastro.2013.10.045
- Stanich PP, Pearlman R. Hereditary or not? Understanding serrated polyposis syndrome. Curr Treat Options Gastroenterol [Internet]. 2019;17(4):692–701. Disponible en: https://doi.org/10.1007/s11938-019-00256-z
- Bonilla CE, Montenegro P, O’Connor JM, et al. Ibero-American consensus review and incorporation of new biomarkers for clinical practice in colorectal cancer. Cancers (Basel) [Internet]. 2023;15(17):4373. Disponible en: https://doi.org/10.3390/cancers15174373
- Abdel Hamid M, Pammer LM, Oberparleiter S, et al. Multidimensional differences of right- and left-sided colorectal cancer and their impact on targeted therapies. NPJ Precis Oncol [Internet]. 2025;9(1):116. Disponible en: https://doi.org/10.1038/s41698-025-00892-y
- Lansom J, Liew I, Ng KS, et al. Right vs. left colorectal cancer: where do we draw the line? Hum Pathol [Internet]. 2024;151:105634. Disponible en: https://doi.org/10.1016/j.humpath.2024.105634
- Gutierrez C, et al. The prevalence and prognosis of microsatellite instability-high/mismatch repair-deficient colorectal adenocarcinomas in the United States. JCO Precis Oncol [Internet]. 2023;7:e2200179. Disponible en: https://doi.org/10.1200/po.22.00179
- Evrard C, Tachon G, Randrian et al. Microsatellite instability: diagnosis, heterogeneity, discordance, and clinical impact in colorectal cancer. Cancers (Basel) [Internet]. 2019;11(10):1567. Disponible en: https://doi.org/10.3390/cancers11101567
- Cervantes A, Candia Montero L, Pentheroudakis G, et al. Metastatic colorectal cancer: ESMO living guidelines, version 1.2, September 2024. Ann Oncol [Internet]. 2023;34(1):10–32. Disponible en: https://www.esmo.org/guidelines/living-guidelines/esmo-living-guideline-metastatic-colorectal-cancer
- National Comprehensive Cancer Network (NCCN). NCCN Clinical Practice Guidelines in Oncology: Colon Cancer. Version 4.2025. June 27, 2025 [Internet]. Plymouth Meeting (PA): NCCN; 2025 [citado 10 sep 2025]. Disponible en: https://www.nccn.org/professionals/physician_gls/pdf/colon.pdf
- Levin-Sparenberg E, Bylsma LC, Lowe K, et al. A systematic literature review and meta-analysis describing the prevalence of KRAS, NRAS, and BRAF gene mutations in metastatic colorectal cancer. Gastroenterology Res [Internet]. 2020;13(5):184–198. Disponible en: https://doi.org/10.14740/gr1167
- Mao C, Wu XY, Yang ZY, et al. Concordant analysis of KRAS, BRAF, PIK3CA mutations, and PTEN expression between primary colorectal cancer and matched metastases. Sci Rep [Internet]. 2015;5:8065. Disponible en: https://doi.org/10.1038/srep08065
- Akkus E, Öksüz NE, Erul E. KRAS G12C inhibitors as monotherapy or in combination for metastatic colorectal cancer: a proportion and comparative meta-analysis of efficacy and toxicity from phase I-II-III trials. Crit Rev Oncol Hematol [Internet]. 2025;211:104741. Disponible en: https://doi.org/10.1016/j.critrevonc.2025.104741
- Trunk A, Braithwaite M, Nevala-Plagemann C, et al. Real-world outcomes of patients with BRAF-mutated metastatic colorectal cancer treated in the United States. J Natl Compr Canc Netw [Internet]. 2022;20(2):144–150. Disponible en: https://doi.org/10.6004/jnccn.2021.7059
- Pietrantonio F, Petrelli F, Coinu A, et al. Predictive role of BRAF mutations in patients with advanced colorectal cancer receiving cetuximab and panitumumab: a meta-analysis. Eur J Cancer [Internet]. 2015;51(5):587–594. Disponible en: https://doi.org/10.1016/j.ejca.2015.01.054
- Elez E, Yoshino T, Shen L, et al. Encorafenib, cetuximab, and mFOLFOX6 in BRAF-mutated colorectal cancer. N Engl J Med [Internet]. 2025;392(24):2425–2437. Disponible en: https://doi.org/10.1056/NEJMoa2501912
- Kopetz S, Grothey A, Yaeger R, et al. Encorafenib, binimetinib, and cetuximab in BRAF V600E-mutated colorectal cancer. N Engl J Med [Internet]. 2019;381(17):1632–1643. Disponible en: https://doi.org/10.1056/NEJMoa1908075
- Tabernero J, Grothey A, Van Cutsem E, et al. Encorafenib plus cetuximab as a new standard of care for previously treated BRAF V600E-mutant metastatic colorectal cancer: updated survival results and subgroup analyses from the BEACON study. J Clin Oncol [Internet]. 2021;39(4):273–284. Disponible en: https://doi.org/10.3410/f.739464054.793584664
- Singh H, Kang A, Bloudek L, et al. Systematic literature review and meta-analysis of HER2 amplification, overexpression, and positivity in colorectal cancer. JNCI Cancer Spectr [Internet]. 2024;8(1):pkad082. Disponible en: https://doi.org/10.1093/jncics/pkad082
- Venturini J, Massaro G, Lavacchi D, et al. The emerging HER2 landscape in colorectal cancer: the key to unveil the future treatment algorithm? Crit Rev Oncol Hematol [Internet]. 2024;204:104515. Disponible en: https://doi.org/10.1016/j.critrevonc.2024.104515
- Germani MM, Borelli B, Hashimoto T, et al. Impact of Human Epidermal Growth Factor Receptor 2 in patients with metastatic colorectal cancer treated with chemotherapy plus bevacizumab or anti-EGFRs: exploratory analysis of eight randomized trials. J Clin Oncol [Internet]. 2025 Sep 4. Disponible en: https://doi.org/10.1200/JCO-25-01003
- Strickler JH, Cercek A, Siena S, et al. Tucatinib plus trastuzumab for chemotherapy-refractory, HER2-positive, RAS wild-type unresectable or metastatic colorectal cancer (MOUNTAINEER): a multicentre, open-label, phase 2 study. Lancet Oncol [Internet]. 2023;24(5):496–508. Disponible en: https://doi.org/10.1016/S1470-2045(23)00150-X
- Yoshino T, Di Bartolomeo M, Raghav K, et al. Final results of DESTINY-CRC01 investigating trastuzumab deruxtecan in patients with HER2-expressing metastatic colorectal cancer. Nat Commun [Internet]. 2023;14(1):3332. Disponible en: https://doi.org/10.1038/s41467-023-38032-4
- Raghav K, Siena S, Takashima A, et al. Trastuzumab deruxtecan in patients with HER2-positive advanced colorectal cancer (DESTINY-CRC02): primary results from a multicentre, randomised, phase 2 trial. Lancet Oncol [Internet]. 2024;25(9):1147–1162. Disponible en: https://doi.org/10.1016/S1470-2045(24)00380-2
- Sartore-Bianchi A, Trusolino L, Martino C, et al. Dual-targeted therapy with trastuzumab and lapatinib in treatment-refractory, KRAS wild-type, HER2-positive metastatic colorectal cancer (HERACLES). Lancet Oncol [Internet]. 2016;17(6):738–746. Disponible en: https://doi.org/10.1016/S1470-2045(16)00150-9
- Sartore-Bianchi A, Lonardi S, Martino C, et al. Pertuzumab and trastuzumab emtansine in patients with HER2-amplified metastatic colorectal cancer: the phase II HERACLES-B trial. ESMO Open [Internet]. 2020;5(5):e000911. Disponible en: https://doi.org/10.1136/esmoopen-2020-000911
- Ambrosini M, Rousseau B, Manca P, et al. Immune checkpoint inhibitors for POLE or POLD1 proofreading-deficient metastatic colorectal cancer. Ann Oncol [Internet]. 2024;35(7):643–655. Disponible en: https://doi.org/10.1016/j.annonc.2024.03.009
- Bourdais R, Rousseau B, Pujals A, et al. Polymerase proofreading domain mutations: new opportunities for immunotherapy in hypermutated colorectal cancer beyond MMR deficiency. Crit Rev Oncol Hematol [Internet]. 2017;113:242–248. Disponible en: https://doi.org/10.1016/j.critrevonc.2017.03.027
- Castellucci E, He T, Goldstein DY, et al. DNA polymerase ε deficiency leading to an ultramutator phenotype: a novel clinically relevant entity. Oncologist [Internet]. 2017;22(5):497–502. Disponible en: https://doi.org/10.1634/theoncologist.2017-0034
- Marques A, Cavaco P, Torre C, et al. Tumor mutational burden in colorectal cancer: implications for treatment. Crit Rev Oncol Hematol [Internet]. 2024;197:104342. Disponible en: https://doi.org/10.1016/j.critrevonc.2024.104342
- Marabelle A, Fakih M, Lopez J, et al. Association of tumour mutational burden with outcomes in patients with advanced solid tumours treated with pembrolizumab (KEYNOTE-158). Lancet Oncol [Internet]. 2020;21(10):1353–1365. Disponible en: https://doi.org/10.1016/S1470-2045(20)30445-9
- Sullo FG, Garinet S, Blons H, et al. Molecular features and clinical actionability of gene fusions in colorectal cancer. Crit Rev Oncol Hematol [Internet]. 2025;208:104656. Disponible en: https://doi.org/10.1016/j.critrevonc.2025.104656
- Cocco E, Scaltriti M, Drilon A. NTRK fusion-positive cancers and TRK inhibitor therapy. Nat Rev Clin Oncol[Internet]. 2018;15(12):731–747. Disponible en: https://doi.org/10.1038/s41571-018-0113-0
- Wang H, Tang R, Jiang L, et al. The role of PIK3CA gene mutations in colorectal cancer and treatment selection. Front Pharmacol [Internet]. 2024;15:1494802. Disponible en: https://doi.org/10.3389/fphar.2024.1494802
- Martling A, Lindberg J, Myrberg IH, et al. Low-dose aspirin to reduce recurrence in colorectal cancer patients with PI3K pathway alterations: ALASCCA trial. J Clin Oncol [Internet]. 2025;43(4 Suppl):LBA125. Disponible en: https://doi.org/10.1200/JCO.2025.43.4_suppl.LBA125
- Lyu X, Cai R, Han B, et al. FGFR1-amplified colorectal cancer: a distinct prognostic subtype. ESMO Open [Internet]. 2025;10(9):105561. Disponible en: https://doi.org/10.1016/j.esmoop.2025.105561
- Lyu X, Cai R, Han B, et al. Comprehensive landscape of FGFR variations in colorectal cancer from ctDNA and tissue analysis. J Clin Oncol [Internet]. 2025;43(4 Suppl):278. Disponible en: https://doi.org/10.1200/JCO.2025.43.4_suppl.278
- Wheless MC, Zemla TJ, Hubbard JM, et al. Phase II study of pemigatinib in metastatic colorectal cancer with FGFR alterations. Oncologist [Internet]. 2025;30(6):oyaf069. Disponible en: https://doi.org/10.1093/oncolo/oyaf069
- Dienstmann R, Vermeulen L, Guinney J, et al. Consensus molecular subtypes and the evolution of precision medicine in colorectal cancer. Nat Rev Cancer [Internet]. 2017;17(2):79–92. Disponible en: https://doi.org/10.1038/nrc.2016.126
- Isella C, Brundu F, Bellomo SE, et al. Cancer-cell intrinsic transcriptional traits define clinically relevant colorectal cancer subtypes. Nat Commun [Internet]. 2017;8:15107. Disponible en: https://doi.org/10.1038/ncomms15107
- Malla SB, Byrne RM, Lafarge MW, et al. Pathway-level subtyping identifies a slow-cycling phenotype in colorectal cancer. Nat Genet [Internet]. 2024;56(3):458–472. Disponible en: https://doi.org/10.1038/s41588-024-01654-5
- Pagès F, Mlecnik B, Marliot F, et al. International validation of the consensus Immunoscore in colon cancer: a prognostic and accuracy study. Lancet [Internet]. 2018;391(10135):2128–2139. Disponible en: https://doi.org/10.1016/S0140-6736(18)30789-X
- Salazar R, Roepman P, Capella G, et al. Gene expression signature to improve prognosis prediction in stage II–III colorectal cancer. J Clin Oncol [Internet]. 2011;29(1):17–24. Disponible en: https://doi.org/10.1200/JCO.2010.30.1077
- You YN, Rustin RB, Sullivan JD. Oncotype DX colon cancer assay for recurrence risk prediction. Surg Oncol[Internet]. 2015;24(2):61–66. Disponible en: https://doi.org/10.1016/j.suronc.2015.02.001
- Niedzwiecki D, Frankel WL, Venook AP, et al. Gene expression signature and recurrence-free interval in stage II colon cancer. J Clin Oncol [Internet]. 2016;34(25):3047–3053. Disponible en: https://doi.org/10.1200/JCO.2015.65.4699
- Mauri G, Vitiello PP, Sogari A, et al. Liquid biopsies to monitor and direct treatment in colorectal cancer. Br J Cancer[Internet]. 2022;127(3):394–407. Disponible en: https://doi.org/10.1038/s41416-022-01769-8
- Malla M, Loree JM, Kasi PM, et al. Using circulating tumor DNA in colorectal cancer. J Clin Oncol [Internet]. 2022;40(24):2846–2857. Disponible en: https://doi.org/10.1200/JCO.21.02615
- Reinert T, Henriksen TV, Christensen E, et al. Analysis of plasma cell-free DNA analysis by ultradeep sequencing in patients with stages I to IIi colorectal cancer. JAMA Oncol [Internet]. 2019;5(8):1124–1131. Disponible en: https://doi.org/10.1001/jamaoncol.2019.0528
- Tarazona N, Gimeno-Valiente F, Gambardella V, et al. Targeted next-generation sequencing of circulating-tumor DNA for tracking minimal residual disease in localized colon cance. Ann Oncol [Internet]. 2019;30(11):1804–1812. Disponible en: https://doi.org/10.1093/annonc/mdz390
- Tie J, Wang Y, Tomasetti C, et al. Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med [Internet]. 2016;8(346):346ra92. Disponible en: https://doi.org/10.1007/s11725-017-0702-6
- Kotani D, Oki E, Nakamura Y, et al. Molecular residual disease and efficacy of adjuvant chemotherapy in patients with colorectal cancer. Nat Med [Internet]. 2023;29(1):127–134. Disponible en: https://doi.org/10.1038/s41591-022-02115-4
- Tie J, Cohen JD, Wang Y, et al. Circulating tumor DNA analyses as markers of recurrence risk and benefit of adjuvant therapy for stage III colon cancer. JAMA Oncol [Internet]. 2019;5(12):1710–1717. Disponible en: https://doi.org/10.1001/jamaoncol.2019.3616
- Tie J, Wang Y, Lo SN, et al. ctDNA-guided adjuvant therapy in stage II colon cancer: DYNAMIC trial. Nat Med[Internet]. 2025;31(9):1509–1518. Disponible en: https://doi.org/10.1038/s41591-025-03579-6
- Patelli G, Lazzari L, Crisafulli G, et al. Clinical utility and future perspectives of liquid biopsy in colorectal cancer. Commun Med [Internet]. 2025;5(1):137. Disponible en: https://doi.org/10.1038/s43856-025-00852-4
- Tao XY, Li QQ, Zeng Y. Clinical application of liquid biopsy in colorectal cancer: detection, prediction, and treatment monitoring Mol Cancer [Internet]. 2024;23(1):145. Disponible en: https://doi.org/10.1186/s12943-024-02063-2
- Parseghian CM, Loree JM, Morris VK, et al. Anti-EGFR-resistant clones decay exponentially after progression: implications for anti-EGFR rechallenge. Ann Oncol [Internet]. 2019;30(2):243–249. Disponible en: https://doi.org/10.1093/annonc/mdy509
- Siravegna G, Mussolin B, Buscarino M, et al. Clonal evolution and resistance to EGFR blockade in the blood of colorectal cancer patients. Nat Med [Internet]. 2015;21(7):795–801. Disponible en: https://doi.org/10.1038/nm.3870
- Montagut C, Dalmases A, Bellosillo B, et al. EGFR extracellular domain mutation conferring cetuximab resistance. Nat Med [Internet]. 2012;18(2):221–223. Disponible en: https://doi.org/10.1038/nm.2609
- Corcoran RB, André T, Atreya CE, et al. Combined BRAF, EGFR and MEK inhibition in patients with BRAF V600E-mutant colorectal cancer. Cancer Discov [Internet]. 2018;8(4):428–443. Disponible en: https://doi.org/10.1158/2159-8290.CD-17-1226
- Martinelli E, Martini G, Famiglietti V, Troiani T, et al. Cetuximab rechallenge plus avelumab in RAS wild-type metastatic colorectal cancer: the phase 2 single-arm clinical CAVE trial. JAMA Oncol [Internet]. 2021;7(10): 1529–1535. Disponible en: https://doi.org/10.1001/jamaoncol.2021.2915
- Cremolini C, Rossini D, Dell’Aquila E, et al. Rechallenge for patients with RAS and BRAF wild-type metastatic colorectal cancer with acquired resistance to first-line cetuximab and irinotecan: a phase 2 single-arm clinical trial. JAMA Oncol [Internet]. 2019;5(5):343–350. Disponible en: https://doi.org/10.1001/jamaoncol.2018.5080
- Parseghian CM, Napolitano S, Loree JM, et al. Mechanisms of resistance to anti-EGFR therapy and rechallenge strategies. Clin Cancer Res [Internet]. 2019;25(23):6899–6908. Disponible en: https://doi.org/10.1158/1078-0432.CCR-19-0823
- Pietrantonio F, Vernieri C, Siravegna G, et al. Heterogeneity of acquired resistance to anti-EGFR monoclonal antibodies. Clin Cancer Res [Internet]. 2017;23(10):2414–2422. Disponible en: https://doi.org/10.1158/1078-0432.CCR-16-1863
- A Phase II Randomized Therapeutic Optimization Trial for Subjects With Refractory Metastatic Colorectal Cancer Using ctDNA: Rapid 1 Trial. ClinicalTrials.gov [Internet]. 2021. Disponible en: https://clinicaltrials.gov/study/NCT04786600
- Circulating Cell-Free Tumor DNA Testing in Guiding Treatment for Patients With Advanced or Metastatic Colorectal Cancer. ClinicalTrials.gov [Internet]. 2019. Disponible en: https://clinicaltrials.gov/study/NCT03844620
- Thierry AR, Mouliere F, El Messaoudi S, et al. Clinical validation of the detection of KRAS and BRAF mutation circulating tumor DNA. Nat Med [Internet]. 2014;20(4):430–435. Disponible en: https://doi.org/10.1038/nm.3511
- Martínez-Castedo B, Camblor DG, Martín-Arana J, et al. Minimal residual disease in colorectal cancer: tumor-informed versus tumor-agnostic approaches: unraveling the optimal strategy. Ann Oncol [Internet]. 2025;36(5):345–357. Disponible en: https://doi.org/10.1016/j.annonc.2024.12.006
- Normanno N, Esposito Abate R, Lambiase M, et al. RAS testing of liquid biopsy correlates with the outcome of metastatic colorectal cancer patients treated with first-line FOLFIRI plus cetuximab in the CAPRI-GOIM trial. Ann Oncol [Internet]. 2018;29(1):112–118. Disponible en: https://doi.org/10.1093/annonc/mdx417
- Parikh AR, Van Seventer EE, Siravegna G, et al. Minimal residual disease detection using plasma-only ctDNA assay in colorectal cancer. Clin Cancer Res [Internet]. 2021;27(20):5586–5594. Disponible en: https://doi.org/10.1158/1078-0432.CCR-21-0410