Hacia una medicina de precisión: biomarcadores moleculares para la detección temprana y el pronóstico en cáncer de cuello uterino
Toward precision medicine: molecular biomarkers for early detection and prognosis in cervical cancer
Cómo citar
Descargar cita

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Mostrar biografía de los autores
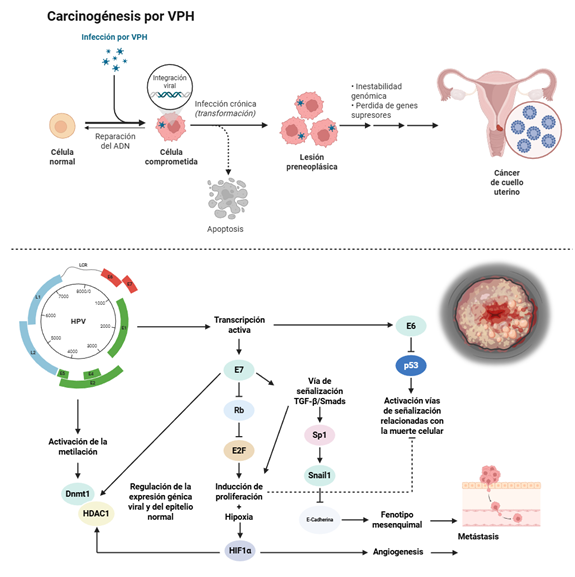
Introducción: el cáncer de cuello uterino (CCU) sigue siendo un desafío de salud pública global, especialmente en países de ingresos bajos y medianos, con baja cobertura de vacunación contra el virus del papiloma humano (VPH) y programas de cribado limitados. Aunque la citología y la detección de ADN de VPH han reducido la mortalidad, sus limitaciones en sensibilidad y especificidad generan sobrediagnóstico y procedimientos innecesarios. En este contexto, los biomarcadores moleculares han surgido como herramientas prometedoras para mejorar la detección temprana, el cribado y la estratificación pronóstica del CCU.
Métodos: se realizó una revisión narrativa de la literatura basada en estudios clínicos y traslacionales que evalúan biomarcadores inmunohistoquímicos, epigenéticos y transcriptómicos asociados con la progresión del CCU.
Resultados: entre los biomarcadores inmunohistoquímicos, p16^INK4a^, Ki67, MCM2 y TOP2A demostraron mejorar la precisión diagnóstica y reducir derivaciones innecesarias. En el ámbito epigenético, la metilación del ADN viral (región L1) y de genes del huésped, como PAX1 y SOX1 se asociaron con lesiones de alto grado. Asimismo, los oncomiRNAs como miR-21 y miR-155 y supresores como miR-126 y miR-143 se relacionaron con la progresión tumoral y el pronóstico, y han mostrado ser útiles en biopsias líquidas, lo que refuerza su potencial como marcadores no invasivos. Los avances en epigenómica y transcriptómica han permitido identificar paneles multibiomarcadores con sensibilidad y especificidad superiores al 80%.
Conclusión: los biomarcadores moleculares representan herramientas prometedoras para un cribado de precisión en CCU. No obstante, la heterogeneidad metodológica exige estudios multicéntricos y de validación prospectiva antes de su implementación clínica.
Visitas del artículo 0 | Visitas PDF 0
Descargas
- Hull R, Mbele M, Makhafola T, et al. Cervical cancer in low and middle‑income countries (Review). Oncology Letters. [Internet] 2020;20(3):2058-2074. Disponible en: https://doi.org/10.3892/ol.2020.11754
- Li Z, Liu P, Yin A, et al. Global landscape of cervical cancer incidence and mortality in 2022 and predictions to 2030: The urgent need to address inequalities in cervical cancer. International Journal of Cancer. [Internet] 2025;157(2):288-297. Disponible en: https://doi.org/10.1002/ijc.35369
- de Martel C, Plummer M, Vignat J, Franceschi S. Worldwide burden of cancer attributable to HPV by site, country and HPV type. International Journal of Cancer. [Internet] 2017;141(4):664-670. Disponible en: https://doi.org/10.1002/ijc.30716
- Bowden SJ, Doulgeraki T, Bouras E, et al. Risk factors for human papillomavirus infection, cervical intraepithelial neoplasia and cervical cancer: an umbrella review and follow-up Mendelian randomisation studies. BMC Medicine. [Internet] 2023;21(1):274. Disponible en: https://doi.org/10.1186/s12916-023-02965-w
- Tekalign T, Teshome M. Prevalence and determinants of late-stage presentation among cervical cancer patients, a systematic review and meta-analysis. Ekwunife OI, ed. PLOS ONE. [Internet] 2022;17(4):e0267571. Disponible en: https://doi.org/10.1371/journal.pone.0267571
- Koliopoulos G, Nyaga VN, Santesso N, et al. Cytology versus HPV testing for cervical cancer screening in the general population. Cochrane Database of Systematic Reviews. [Internet] 2017;2018(7). Disponible en: https://doi.org/10.1002/14651858.cd008587.pub2
- Haedicke J, Iftner T. Human papillomaviruses and cancer. Radiotherapy and Oncology. [Internet] 2013;108(3):397-402. Disponible en: https://doi.org/10.1016/j.radonc.2013.06.004
- Volkova L V., Pashov AI, Omelchuk NN. Cervical Carcinoma: Oncobiology and Biomarkers. International Journal of Molecular Sciences. [Internet] 2021;22(22):12571. Disponible en: https://doi.org/10.3390/ijms222212571
- de Sanjose S, Quint WGV, Alemany L, et al. Human papillomavirus genotype attribution in invasive cervical cancer: a retrospective cross-sectional worldwide study. Lancet Oncol. [Internet] 2010;11(11):1048-1056. Disponible en: https://doi.org/10.1016/S1470-2045(10)70230-8
- Senapati R, Senapati NN, Dwibedi B. Molecular mechanisms of HPV mediated neoplastic progression. Infectious Agents and Cancer. [Internet] 2016;11(1):59. Disponible en: https://doi.org/10.1186/s13027-016-0107-4
- Chu D, Liu T, Yao Y. Implications of viral infections and oncogenesis in uterine cervical carcinoma etiology and pathogenesis. Frontiers in Microbiology. [Internet] 2023;14. Disponible en: https://doi.org/10.3389/fmicb.2023.1194431
- zur Hausen H. Papillomaviruses and cancer: from basic studies to clinical application. Nature Reviews Cancer. [Internet] 2002;2(5):342-350. Disponible en: https://doi.org/10.1038/nrc798
- Fernandes JV, De Medeiros Fernandes TAA, De Azevedo JCV, et al. Link between chronic inflammation and human papillomavirus-induced carcinogenesis (Review). Oncology Letters. [Internet] 2015;9(3):1015-1026. Disponible en: https://doi.org/10.3892/ol.2015.2884
- Bhattacharjee R, Das SS, Biswal SS, et al. Mechanistic role of HPV-associated early proteins in cervical cancer: Molecular pathways and targeted therapeutic strategies. Critical Reviews in Oncology/Hematology. [Internet] 2022;174:103675. Disponible en: https://doi.org/10.1016/j.critrevonc.2022.103675
- Parvizi M, Vaezi M, Jeddi F, et al. The role and diagnostic value of deregulated miRNAs in cervical cancer. Discover Oncology. [Internet] 2025;16(1):922. Disponible en: https://doi.org/10.1007/s12672-025-02744-4
- Molina MA, Carosi Diatricch L, Castany Quintana M, Melchers WJ, Andralojc KM. Cervical cancer risk profiling: molecular biomarkers predicting the outcome of hrHPV infection. Expert Review of Molecular Diagnostics. [Internet] 2020;20(11):1099-1120. Disponible en: https://doi.org/10.1080/14737159.2020.1835472
- Lucksom PG, Sherpa ML, Pradhan A, Lal S, Gupta C. Advances in HPV Screening Tests for Cervical Cancer—A Review. The Journal of Obstetrics and Gynecology of India. [Internet] 2022;72(1):13-18. Disponible en: https://doi.org/10.1007/s13224-021-01569-9
- The ASCUS-LSIL Triage Study (ALTS)* Group. Results of a randomized trial on the management of cytology interpretations of atypical squamous cells of undetermined significance. American Journal of Obstetrics and Gynecology. [Internet] 2003;188(6):1383-1392. Disponible en: https://doi.org/10.1016/s0002-9378(03)00418-6
- Gilham C, Sargent A, Kitchener HC, Peto J. HPV testing compared with routine cytology in cervical screening: long-term follow-up of ARTISTIC RCT. Health Technology Assessment. [Internet] 2019;23(28):1-44. Disponible en: https://doi.org/10.3310/hta23280
- Ogilvie GS, van Niekerk D, Krajden M, et al. Effect of Screening With Primary Cervical HPV Testing vs Cytology Testing on High-grade Cervical Intraepithelial Neoplasia at 48 Months: The HPV FOCAL Randomized Clinical Trial. Obstetrical & Gynecological Survey. [Internet] 2018;73(11):632-634. Disponible en: https://doi.org/10.1097/ogx.0000000000000608
- Siebers AG, Arbyn M, Melchers WJG, et al. Effectiveness of two strategies to follow-up ASC-US and LSIL screening results in The Netherlands using repeat cytology with or without additional hrHPV testing: a retrospective cohort study. Cancer Causes & Control. [Internet] 2014;25(9):1141-1149. Disponible en: https://doi.org/10.1007/s10552-014-0414-2
- Maver PJ, Poljak M. Primary HPV-based cervical cancer screening in Europe: implementation status, challenges, and future plans. Clinical microbiology and infection : the official publication of the European Society of Clinical Microbiology and Infectious Diseases. [Internet] 2020;26(5):579-583. Disponible en: https://doi.org/10.1016/j.cmi.2019.09.006
- Wright TC, Stoler MH, Behrens CM, Apple R, Derion T, Wright TL. The ATHENA human papillomavirus study: design, methods, and baseline results. American Journal of Obstetrics and Gynecology. [Internet] 2012;206(1):46.e1-46.e11. Disponible en: https://doi.org/10.1016/j.ajog.2011.07.024
- Dunne EF, Unger ER, Sternberg M, et al. Prevalence of HPV Infection Among Females in the United States. JAMA. [Internet] 2007;297(8):813. Disponible en: https://doi.org/10.1001/jama.297.8.813
- Kang L-N, Jeronimo J, Qiao Y-L, et al. Optimal Positive Cutoff Points for careHPV Testing of Clinician- and Self-Collected Specimens in Primary Cervical Cancer Screening: an Analysis from Rural China. Tang Y-W, ed. Journal of Clinical Microbiology. [Internet] 2014;52(6):1954-1961. Disponible en: https://doi.org/10.1128/jcm.03432-13
- Kocken M, Uijterwaal MH, de Vries ALM, et al. High-risk human papillomavirus testing versus cytology in predicting post-treatment disease in women treated for high-grade cervical disease: A systematic review and meta-analysis. Gynecologic Oncology. [Internet] 2012;125(2):500-507. Disponible en: https://doi.org/10.1016/j.ygyno.2012.01.015
- Wentzensen N, Schiffman M, Palmer T, Arbyn M. Triage of HPV positive women in cervical cancer screening. Journal of Clinical Virology. [Internet] 2016;76:S49-S55. Disponible en: https://doi.org/10.1016/j.jcv.2015.11.015
- Constandinou-Williams C, Collins SI, Roberts S, Young LS, Woodman CBJ, Murray PG. Is Human Papillomavirus Viral Load a Clinically Useful Predictive Marker? A Longitudinal Study. Cancer Epidemiology, Biomarkers & Prevention. [Internet] 2010;19(3):832-837. Disponible en: https://doi.org/10.1158/1055-9965.epi-09-0838
- Kim J, Kim BK, Jeon D, et al. Type-Specific Viral Load and Physical State of HPV Type 16, 18, and 58 as Diagnostic Biomarkers for High-Grade Squamous Intraepithelial Lesions or Cervical Cancer. Cancer Research and Treatment. [Internet] 2020;52(2):396-405. Disponible en: https://doi.org/10.4143/crt.2019.152
- Cao M, Wang Y, Wang D, et al. Increased High-Risk Human Papillomavirus Viral Load Is Associated With Immunosuppressed Microenvironment and Predicts a Worse Long-Term Survival in Cervical Cancer Patients. American Journal of Clinical Pathology. [Internet] 2020;153(4):502-512. Disponible en: https://doi.org/10.1093/ajcp/aqz186
- van den Heuvel CNAM, Loopik DL, Ebisch RMF, et al. RNA-based high-risk HPV genotyping and identification of high-risk HPV transcriptional activity in cervical tissues. Modern Pathology. [Internet] 2020;33(4):748-757. Disponible en: https://doi.org/10.1038/s41379-019-0369-7
- Benevolo M, Vocaturo A, Caraceni D, et al. Sensitivity, Specificity, and Clinical Value of Human Papillomavirus (HPV) E6/E7 mRNA Assay as a Triage Test for Cervical Cytology and HPV DNA Test. Journal of Clinical Microbiology. [Internet] 2011;49(7):2643-2650. Disponible en: https://doi.org/10.1128/jcm.02570-10
- Cattani P, Zannoni GF, Ricci C, et al. Clinical Performance of Human Papillomavirus E6 and E7 mRNA Testing for High-Grade Lesions of the Cervix. Journal of Clinical Microbiology. [Internet] 2009;47(12):3895-3901. Disponible en: https://doi.org/10.1128/jcm.01275-09
- Ge Y, Christensen P, Luna E, Armylagos D, Schwartz MR, Mody DR. Performance of A ptima and C obas HPV testing platforms in detecting high‐grade cervical dysplasia and cancer. Cancer Cytopathology. [Internet] 2017;125(8):652-657. Disponible en: https://doi.org/10.1002/cncy.21875
- Macedo ACL, Gonçalves JCN, Bavaresco DV, Grande AJ, Chiaramonte Silva N, Rosa MI. Accuracy of mRNA HPV Tests for Triage of Precursor Lesions and Cervical Cancer: A Systematic Review and Meta-Analysis. Journal of Oncology. [Internet] 2019;2019:1-14. Disponible en: https://doi.org/10.1155/2019/6935030
- Bruno MT, Ferrara M, Fava V, Barrasso G, Panella MM. A prospective study of women with ASCUS or LSIL pap smears at baseline and HPV E6/E7 mRNA positive: a 3-year follow-up. Epidemiology and Infection. [Internet] 2018;146(5):612-618. Disponible en: https://doi.org/10.1017/s0950268818000250
- Duvlis S, Popovska-Jankovic K, Arsova ZS, Memeti S, Popeska Z, Plaseska-Karanfilska D. HPV E6/E7 mRNA versus HPV DNA biomarker in cervical cancer screening of a group of Macedonian women. Journal of Medical Virology. [Internet] 2015;87(9):1578-1586. Disponible en: https://doi.org/10.1002/jmv.24199
- Lindroth Y, Borgfeldt C, Thorn G, Bodelsson G, Forslund O. Population-based primary HPV mRNA cervical screening compared with cytology screening. Preventive Medicine. [Internet] 2019;124:61-66. Disponible en: https://doi.org/10.1016/j.ypmed.2019.04.021
- Sørbye SW, Fismen S, Gutteberg TJ, Mortensen ES, Skjeldestad FE. Primary cervical cancer screening with an HPV mRNA test: a prospective cohort study. BMJ Open. [Internet] 2016;6(8):e011981. Disponible en: https://doi.org/10.1136/bmjopen-2016-011981
- Sørbye S, Falang BM, Antonsen M, Mortensen E. Genotype-Specific HPV mRNA Triage Improves CIN2+ Detection Efficiency Compared to Cytology: A Population-Based Study of HPV DNA-Positive Women. Pathogens. [Internet] 2025;14(8):749. Disponible en: https://doi.org/10.3390/pathogens14080749
- Hsu Y-W, Huang R-L, Su P-H, et al. Genotype-specific methylation of HPV in cervical intraepithelial neoplasia. Journal of Gynecologic Oncology. [Internet] 2017;28(4):e56. Disponible en: https://doi.org/10.3802/jgo.2017.28.e56
- Vinokurova S, von Knebel Doeberitz M. Differential Methylation of the HPV 16 Upstream Regulatory Region during Epithelial Differentiation and Neoplastic Transformation. Ramqvist T, ed. PLoS ONE. [Internet] 2011;6(9):e24451. Disponible en: https://doi.org/10.1371/journal.pone.0024451
- Kalantari M, Osann K, Calleja-Macias IE, et al. Methylation of human papillomavirus 16, 18, 31, and 45 L2 and L1 genes and the cellular DAPK gene: Considerations for use as biomarkers of the progression of cervical neoplasia. Virology. [Internet] 2014;448:314-321. Disponible en: https://doi.org/10.1016/j.virol.2013.10.032
- Wentzensen N, Sun C, Ghosh A, et al. Methylation of HPV18, HPV31, and HPV45 genomes and cervical intraepithelial neoplasia grade 3. Journal of the National Cancer Institute. [Internet] 2012;104(22):1738-1749. Disponible en: https://doi.org/10.1093/jnci/djs425
- Wentzensen N, Sun C, Ghosh A, et al. Methylation of HPV18, HPV31, and HPV45 Genomes and Cervical Intraepithelial Neoplasia Grade 3. JNCI: Journal of the National Cancer Institute. [Internet] 2012;104(22):1738-1749. Disponible en: https://doi.org/10.1093/jnci/djs425
- Carozzi F, Gillio-Tos A, Confortini M, et al. Risk of high-grade cervical intraepithelial neoplasia during follow-up in HPV-positive women according to baseline p16-INK4A results: a prospective analysis of a nested substudy of the NTCC randomised controlled trial. The Lancet Oncology. [Internet] 2013;14(2):168-176. Disponible en: https://doi.org/10.1016/s1470-2045(12)70529-6
- Molina MA, Carosi Diatricch L, Castany Quintana M, Melchers WJG, Andralojc KM. Cervical cancer risk profiling: molecular biomarkers predicting the outcome of hrHPV infection. Expert Review of Molecular Diagnostics. [Internet] 2020;20(11):1099-1120. Disponible en: https://doi.org/10.1080/14737159.2020.1835472
- Liu Y, Wen L, Jiang L, Xu J, Meng Y, Yang W. Immunocytochemical staining for p16, Ki-67, and MCM2 in the detection of cervical lesions and cancer: a prospective observational study. Translational Cancer Research. [Internet] 2025;14(5):3201-3211. Disponible en: https://doi.org/10.21037/tcr-2025-802
- Del Moral-Hernández O, Hernández-Sotelo D, Alarcón-Romero L del C, et al. TOP2A/MCM2, p16INK4a, and cyclin E1 expression in liquid-based cytology: a biomarkers panel for progression risk of cervical premalignant lesions. BMC Cancer. [Internet] 2021;21(1):39. Disponible en: https://doi.org/10.1186/s12885-020-07740-1
- Qiu S, Wang Q, Jiang H, Feng L. Immunohistochemistry staining of Eag1 and p16/Ki-67 can help improve the management of patients with cervical intraepithelial Neoplasia after cold knife conversion. Diagnostic Pathology. [Internet] 2024;19(1):97. Disponible en: https://doi.org/10.1186/s13000-024-01523-z
- Usta ZT, Yilmaz ZS, Ersoz S, Mungan SA, Cobanoglu U, Guven S. Comparison of ProExC and p16ink4a Biological Markers in Lesional Smears With the Immunocytochemical Method and Relationship With Human Papillomavirus in Liquid-based Cervicovaginal Specimens. Journal of Cytology. [Internet] 2025;42(1):20-26. Disponible en: https://doi.org/10.4103/joc.joc_57_24
- Damgaard R, Jenkins D, Stoler MH, et al. p16 INK4a and HPV E4 immunohistochemistry for the prediction of regression of cervical intraepithelial neoplasia grade 2—A historical cohort study. International Journal of Cancer. [Internet] 2025;157(7):1294-1303. Disponible en: https://doi.org/10.1002/ijc.35469
- Verlaat W, Van Leeuwen RW, Novianti PW, et al. Host-cell DNA methylation patterns during high-risk HPV-induced carcinogenesis reveal a heterogeneous nature of cervical pre-cancer. Epigenetics. [Internet] 2018;13(7):769-778. Disponible en: https://doi.org/10.1080/15592294.2018.1507197
- Zhou L, Qiu Q, Zhou Q, et al. Long-read sequencing unveils high-resolution HPV integration and its oncogenic progression in cervical cancer. Nature Communications. [Internet] 2022;13(1):2563. Disponible en: https://doi.org/10.1038/s41467-022-30190-1
- De Strooper LMA, Meijer CJLM, Berkhof J, et al. Methylation Analysis of the FAM19A4 Gene in Cervical Scrapes Is Highly Efficient in Detecting Cervical Carcinomas and Advanced CIN2/3 Lesions. Cancer Prevention Research. [Internet] 2014;7(12):1251-1257. Disponible en: https://doi.org/10.1158/1940-6207.capr-14-0237
- Kocsis A, Takács T, Jeney C, et al. Performance of a new HPV and biomarker assay in the management of hrHPV positive women: Subanalysis of the ongoing multicenter TRACE clinical trial ( n > 6,000) to evaluate POU4F3 methylation as a potential biomarker of cervical precancer and cancer. International Journal of Cancer. [Internet] 2017;140(5):1119-1133. Disponible en: https://doi.org/10.1002/ijc.30534
- Vasiljević N, Scibior-Bentkowska D, Brentnall AR, Cuzick J, Lorincz AT. Credentialing of DNA methylation assays for human genes as diagnostic biomarkers of cervical intraepithelial neoplasia in high-risk HPV positive women. Gynecologic Oncology. [Internet] 2014;132(3):709-714. Disponible en: https://doi.org/10.1016/j.ygyno.2014.02.001
- Li N, He Y, Mi P, Hu Y. ZNF582 methylation as a potential biomarker to predict cervical intraepithelial neoplasia type III/worse. Medicine. [Internet] 2019;98(6):e14297. Disponible en: https://doi.org/10.1097/md.0000000000014297
- Kan Y-Y, Liou Y-L, Wang H-J, et al. PAX1 Methylation as a Potential Biomarker for Cervical Cancer Screening. International Journal of Gynecological Cancer. [Internet] 2014;24(5):928-934. Disponible en: https://doi.org/10.1097/igc.0000000000000155
- Lorincz AT. Virtues and Weaknesses of DNA Methylation as a Test for Cervical Cancer Prevention. Acta Cytologica. [Internet] 2016;60(6):501-512. Disponible en: https://doi.org/10.1159/000450595
- Pun PB, Liao Y-P, Su P-H, et al. Triage of high-risk human papillomavirus-positive women by methylated POU4F3. Clinical Epigenetics. [Internet] 2015;7(1):85. Disponible en: https://doi.org/10.1186/s13148-015-0122-0
- Chang C-L, Ho S-C, Su Y-F, et al. DNA methylation marker for the triage of hrHPV positive women in cervical cancer screening: Real-world evidence in Taiwan. Gynecologic Oncology. [Internet] 2021;161(2):429-435. Disponible en: https://doi.org/10.1016/j.ygyno.2021.02.011
- Overmeer RM, Louwers JA, Meijer CJLM, et al. Combined CADM1 and MAL promoter methylation analysis to detect (pre‐)malignant cervical lesions in high‐risk HPV‐positive women. International Journal of Cancer. [Internet] 2011;129(9):2218-2225. Disponible en: https://doi.org/10.1002/ijc.25890
- Bonde J, Floore A, Ejegod D, et al. Methylation markers FAM19A4 and miR124 ‐2 as triage strategy for primary human papillomavirus screen positive women: A large European multicenter study. International Journal of Cancer. [Internet] 2021;148(2):396-405. Disponible en: https://doi.org/10.1002/ijc.33320
- Shi L, Yang X, He L, et al. Promoter hypermethylation analysis of host genes in cervical intraepithelial neoplasia and cervical cancers on histological cervical specimens. BMC Cancer. [Internet] 2023;23(1):168. Disponible en: https://doi.org/10.1186/s12885-023-10628-5
- Zhang L, Zhao X, Hu S, et al. Triage performance and predictive value of the human gene methylation panel among women positive on self‐collected HPV test: Results from a prospective cohort study. International Journal of Cancer. [Internet] 2022;151(6):878-887. Disponible en: https://doi.org/10.1002/ijc.34041
- Vink FJ, Meijer CJLM, Clifford GM, et al. FAM19A4/miR124‐2 methylation in invasive cervical cancer: A retrospective cross‐sectional worldwide study. International Journal of Cancer. [Internet] 2020;147(4):1215-1221. Disponible en: https://doi.org/10.1002/ijc.32614
- Kremer WW, Dick S, Heideman DAM, et al. Clinical Regression of High-Grade Cervical Intraepithelial Neoplasia Is Associated With Absence of FAM19A4/miR124-2 DNA Methylation (CONCERVE Study). Journal of Clinical Oncology. [Internet] 2022;40(26):3037-3046. Disponible en: https://doi.org/10.1200/jco.21.02433
- Zhang L, Yu J, Huang W, Zhang H, Xu J, Cai H. A Sensitive and Simplified Classifier of Cervical Lesions Based on a Methylation-Specific PCR Assay: A Chinese Cohort Study. Cancer Management and Research. [Internet] 2020; 12:2567-2576. Disponible en: https://doi.org/10.2147/cmar.s246103
- Lai H, Ou Y, Chen T, et al. PAX 1 / SOX 1 DNA methylation and cervical neoplasia detection: a Taiwanese Gynecologic Oncology Group ( TGOG ) study. Cancer Medicine. [Internet] 2014;3(4):1062-1074. Disponible en: https://doi.org/10.1002/cam4.253
- de Waard J, Bhattacharya A, de Boer MT, et al. Methylation Analysis to Detect CIN3+ in High-Risk Human Papillomavirus-Positive Self-Samples From the Population-Based Cervical Cancer Screening Program. Modern Pathology. [Internet] 2024;37(8):100528. Disponible en: https://doi.org/10.1016/j.modpat.2024.100528
- Bowden SJ, Bodinier B, Paraskevaidi M, et al. DNA methylation signatures of cervical pre‐invasive and invasive disease: An epigenome‐wide association study. International Journal of Cancer. [Internet] 2025;157(2):305-316. Disponible en: https://doi.org/10.1002/ijc.35406
- El‐Zein M, Cheishvili D, Gotlieb W, et al. Genome‐wide DNA methylation profiling identifies two novel genes in cervical neoplasia. International Journal of Cancer. [Internet] 2020;147(5):1264-1274. Disponible en: https://doi.org/10.1002/ijc.32880
- Boers A, Wang R, van Leeuwen RW, et al. Discovery of new methylation markers to improve screening for cervical intraepithelial neoplasia grade 2/3. Clinical Epigenetics. [Internet] 2016;8(1):29. Disponible en: https://doi.org/10.1186/s13148-016-0196-3
- de Waard J, Bhattacharya A, de Boer MT, et al. Identification of a methylation panel as an alternative triage to detect CIN3+ in hrHPV-positive self-samples from the population-based cervical cancer screening programme. Clinical Epigenetics. [Internet] 2023;15(1):103. d Disponible en: https://doi.org/10.1186/s13148-023-01517-6
- Fackler MJ, Pleas M, Li Y, et al. Discovery and technical validation of high-performance methylated DNA markers for the detection of cervical lesions at risk of malignant progression in low- and middle-income countries. Clinical Epigenetics. [Internet] 2024;16(1):56. Disponible en: https://doi.org/10.1186/s13148-024-01669-z
- Rahimi-Moghaddam A, Ghorbanmehr N, Gharbi S, Nili F, Korsching E. Interplay of miR-542, miR-126, miR-143 and miR-26b with PI3K-Akt is a Diagnostic Signal and Putative Regulatory Target in HPV-Positive Cervical Cancer. Biochemical Genetics. [Internet] 2025;63(3):2760-2780. Disponible en: https://doi.org/10.1007/s10528-024-10837-y
- Yang Y, Song K, Chang H, Chen L. Decreased expression of microRNA-126 is associated with poor prognosis in patients with cervical cancer. Diagnostic Pathology. [Internet] 2014;9(1):220. Disponible en: https://doi.org/10.1186/s13000-014-0220-x
- Shakeri A, Ghanbari M, Tasbandi A, Sahebkar A. Regulation of microRNA ‐21 expression by natural products in cancer. Phytotherapy Research. [Internet] 2021;35(7):3732-3746. Disponible en: https://doi.org/10.1002/ptr.7069
- Knerr S, Guo B, Mittendorf KF, et al. Risk‐reducing surgery in unaffected individuals receiving cancer genetic testing in an integrated health care system. Cancer. [Internet] 2022;128(16):3090-3098. Disponible en: https://doi.org/10.1002/cncr.34349
- Gao D, Zhang Y, Zhu M, Liu S, Wang X. miRNA Expression Profiles of HPV-Infected Patients with Cervical Cancer in the Uyghur Population in China. Tornesello ML, ed. PLOS ONE. [Internet] 2016;11(10):e0164701. Disponible en: https://doi.org/10.1371/journal.pone.0164701
- Du S, Zhao Y, Lv C, Wei M, Gao Z, Meng X. Applying Serum Proteins and MicroRNA as Novel Biomarkers for Early-Stage Cervical Cancer Detection. Scientific Reports. [Internet] 2020;10(1):9033. Disponible en: https://doi.org/10.1038/s41598-020-65850-z
- Kotani K, Iwata A, Kukimoto I, et al. Nomogram for predicted probability of cervical cancer and its precursor lesions using miRNA in cervical mucus, HPV genotype and age. Scientific Reports. [Internet] 2022;12(1):16231. Disponible en: https://doi.org/10.1038/s41598-022-19722-3
- Okoye JO, Ngokere AA, Onyenekwe CC, Erinle CA. Comparable expression of miR-let-7b, miR-21, miR-182, miR-145, and p53 in serum and cervical cells: Diagnostic implications for early detection of cervical lesions. International journal of health sciences. [Internet] 2019;13(4):29-38. Disponible en: http://www.ncbi.nlm.nih.gov/pubmed/31341453.
- Ning R, Meng S, Wang L, et al. 6 Circulating miRNAs can be used as Non-invasive Biomarkers for the Detection of Cervical Lesions. Journal of Cancer. [Internet] 2021;12(17):5106-5113. Disponible en: https://doi.org/10.7150/jca.51141
- Qi Y, Lai Y-L, Shen P-C, et al. Identification and validation of a miRNA-based prognostic signature for cervical cancer through an integrated bioinformatics approach. Scientific Reports. [Internet] 2020;10(1):22270. Disponible en: https://doi.org/10.1038/s41598-020-79337-4
- Zhang J, Li Y, Ren Y, Li J, Han H, Yan P. miR-214-5p increases the radiosensitivity of cervical cancer by targeting ROCK1 expression. Advances in Clinical and Experimental Medicine. [Internet] 2023;33(3):247-259. Disponible en: https://doi.org/10.17219/acem/166673