Relación de la ancestría genética en las alteraciones moleculares en distintos modelos de cáncer
Relationship of genetic ancestry to molecular alterations in different cancer models
Cómo citar
Descargar cita

Esta obra está bajo una licencia internacional Creative Commons Atribución-NoComercial-SinDerivadas 4.0.
Mostrar biografía de los autores
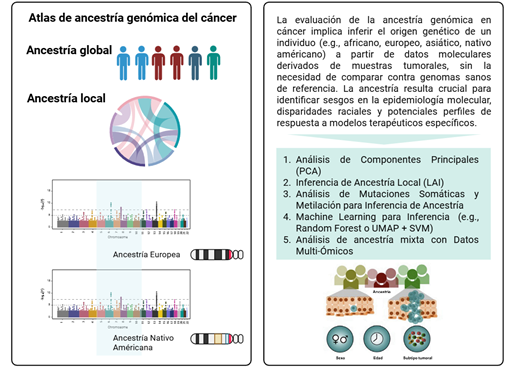
Introducción: las disparidades en cáncer se han atribuido principalmente a determinantes sociales y factores del estilo de vida, sin embargo, evidencia reciente sugiere que la ancestría genética también puede influir en la biología tumoral. Comprender esta relación es clave para avanzar hacia una medicina de precisión más equitativa.
Métodos: se realizó una revisión de la literatura mediante PubMed sin restricciones temporales. Se incluyeron estudios originales que evaluaran la asociación entre la ancestría genética, inferida a partir de marcadores genómicos, y alteraciones moleculares en distintos tipos de cáncer, excluyendo aquellos basados únicamente en autoidentificación étnica.
Resultados: cuarenta estudios cumplieron los criterios de inclusión. Los cánceres más representados fueron mama, pulmón, colon, próstata y neoplasias hematológicas. En cáncer de mama, la ancestría africana se asoció con mutaciones en TP53 y activación de las vías NFκB y EGFR, mientras que la ancestría europea se asoció con mutaciones en PIK3CA y PTEN. En cáncer de pulmón, la ancestría asiática mostró alta prevalencia de mutaciones en EGFR y fusiones de ALK. En cáncer colorrectal, la ancestría africana se asoció con alteraciones en KRAS y APC, y la europea con BRAF y CHEK2. Además, se identificaron asociaciones específicas en cáncer de próstata y leucemia linfoblástica aguda.
Conclusión: la evidencia disponible respalda que la ancestría genética influye en los perfiles moleculares tumorales y puede contribuir a explicar las disparidades en cáncer. Sin embargo, aún persiste una subrepresentación de grupos con alta ancestría nativa americana, como la latinoamericana, lo que limita la equidad en la medicina de precisión.
Visitas del artículo 0 | Visitas PDF 0
Descargas
- Bray F, Laversanne M, Sung H, Ferlay J, Siegel RL, Soerjomataram I, et al. Global cancer statistics 2022: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin [Internet]. 2024;74:229–63. Disponible en: https://doi.org/10.3322/caac.21834
- Elhawary NA, Ekram SN, Sembawa HA, Tashkandi E, Bannani S, Azher ZA, et al. Descriptive epidemiology of female breast cancer around the world: incidence, mortality, and sociodemographic risks and disparities. Int J Environ Health Res [Internet]. 2025;35:3648–62. Disponible en: https://doi.org/10.1080/09603123.2025.2492826
- Benitez Fuentes JD, Morgan E, de Luna Aguilar A, Mafra A, Shah R, Giusti F, et al. Global Stage Distribution of Breast Cancer at Diagnosis. JAMA Oncol [Internet]. 2024;10:71. Disponible en: https://doi.org/10.1001/jamaoncol.2023.4837
- Larsen K, Rydz E, Peters CE. Inequalities in Environmental Cancer Risk and Carcinogen Exposures: A Scoping Review. International Journal of Environmental Research and Public Health 2023, Vol 20, Page 5718 [Internet]. 2023 [citado el 2 de septiembre de 2025;20:5718. Disponible en: https://doi.org/10.3390/ijerph20095718
- Vieira VM, Vopham T, Bertrand KA, James P, Dupré N, Tamimi RM, et al. Contribution of socioeconomic and environmental factors to geographic disparities in breast cancer risk in the Nurses’ Health Study II. Environmental Epidemiology [Internet]. 2020;4(1):p e080. Disponible en: https://doi.org/10.1097/EE9.0000000000000080
- Yuan J, Hu Z, Mahal BA, Zhao SD, Kensler KH, Pi J, et al. Integrated Analysis of Genetic Ancestry and Genomic Alterations across Cancers. Cancer Cell [Internet]. 2018;34:549-560.e9. Disponible en: https://doi.org/10.1016/j.ccell.2018.08.019
- Batai K, Hooker S, Kittles RA. Leveraging genetic ancestry to study health disparities. Am J Phys Anthropol [Internet]. 2021;175:363–75. Disponible en: https://doi.org/10.1002/ajpa.24144
- Mersha TB, Abebe T. Self-reported race/ethnicity in the age of genomic research: Its potential impact on understanding health disparities. Hum Genomics [Internet]. 2015;9:1–15. Disponible en: https://doi.org/10.1186/s40246-014-0023-x
- Alexander DH, Lange K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics [Internet]. 2011;12:1–6. Disponible en: https://doi.org/10.1186/1471-2105-12-246
- Johnson R, Ding Y, Venkateswaran V, Bhattacharya A, Boulier K, Chiu A, et al. Leveraging genomic diversity for discovery in an electronic health record linked biobank: the UCLA ATLAS Community Health Initiative. Genome Med [Internet]. 2022;14:1–23. Disponible en: https://doi.org/10.1186/s13073-022-01128-5
- Lee KK, Rishishwar L, Ban D, Nagar SD, Mariño-Ramírez L, McDonald JF, et al. Association of Genetic Ancestry and Molecular Signatures with Cancer Survival Disparities: A Pan-Cancer Analysis. Cancer Res [Internet]. 2022;82:1222–33. Disponible en: https://doi.org/10.1158/0008-5472.CAN-21-2105
- Carrot-Zhang J, Chambwe N, Damrauer JS, Knijnenburg TA, Robertson AG, Yau C, et al. Comprehensive Analysis of Genetic Ancestry and Its Molecular Correlates in Cancer. Cancer Cell [Internet]. 2020;37(5):639-654.e6. Disponible en: https://doi.org/10.1016/j.ccell.2020.04.012
- Spratt DE, Chan T, Waldron L, Speers C, Feng FY, Ogunwobi O, et al. Racial/Ethnic Disparities in Genomic Sequencing. JAMA Oncol [Internet]. 2016;2:1070–4. Disponible en: https://doi.org/10.1001/jamaoncol.2016.1854
- Atutornu J, Milne R, Costa A, Patch C, Middleton A. Towards equitable and trustworthy genomics research. EBioMedicine [Internet]. 2022;76:103879. Disponible en: https://doi.org/10.1016/j.ebiom.2022.103879
- Oak N, Cherniack AD, Mashl RJ, Carrot-Zhang J, Chambwe N, Damrauer JS, et al. Ancestry-specific predisposing germline variants in cancer. Genome Med [Internet]. 2020;12:1–15. Disponible en: https://doi.org/10.1186/s13073-020-00744-3
- Matejcic M, Teer JK, Hoehn HJ, Diaz DB, Shankar K, Gong J, et al. Colorectal Tumors in Diverse Patient Populations Feature a Spectrum of Somatic Mutational Profiles. Cancer Res [Internet]. 2025;85(10):1928–44. Disponible en: https://doi.org/10.1158/0008-5472.CAN-24-0747
- Yuan J, Kensler KH, Hu Z, Zhang Y, Zhang T, Jiang J, et al. Integrative comparison of the genomic and transcriptomic landscape between prostate cancer patients of predominantly African or European genetic ancestry. PLoS Genet [Internet]. 2020;16:e1008641. Disponible en: https://doi.org/10.1371/journal.pgen.1008641
- Perou CM, Sørile T, Eisen MB, Van De Rijn M, Jeffrey SS, Ress CA, et al. Molecular portraits of human breast tumours. Nature [Internet]. 2000;406:747–52. Disponible en: https://doi.org/10.1038/35021093
- Eroles P, Bosch A, Alejandro Pérez-Fidalgo J, Lluch A. Molecular biology in breast cancer: Intrinsic subtypes and signaling pathways. Cancer Treat Rev [Internet]. 2012;38:698–707. Disponible en: https://doi.org/10.1016/j.ctrv.2011.11.005
- Gnant M, Thomssen C, Harbeck N. St. Gallen/Vienna 2015: A Brief Summary of the Consensus Discussion. Breast Care [Internet]. 2015;10:124–30. Disponible en: https://doi.org/10.1159/000430488
- Huo D, Hu H, Rhie SK, Gamazon ER, Cherniack AD, Liu J, et al. Comparison of Breast Cancer Molecular Features and Survival by African and European Ancestry in The Cancer Genome Atlas. JAMA Oncol [Internet]. 2017;3:1654. Disponible en: https://doi.org/10.1001/jamaoncol.2017.0595
- Dietze EC, Sistrunk C, Miranda-Carboni G, O’Regan R, Seewaldt VL. Triple-negative breast cancer in African-American women: disparities versus biology. Nat Rev Cancer [Internet]. 2015;15:248–54. Disponible en: https://doi.org/10.1038/nrc3896
- Miyashita M, Bell JSK, Wenric S, Karaesmen E, Rhead B, Kase M, et al. Molecular profiling of a real-world breast cancer cohort with genetically inferred ancestries reveals actionable tumor biology differences between European ancestry and African ancestry patient populations. Breast Cancer Research [Internet]. 2023;25:1–13. Disponible en: https://doi.org/10.1186/s13058-023-01627-2
- Davis M, Martini R, Newman L, Elemento O, White J, Verma A, et al. Identification of Distinct Heterogenic Subtypes and Molecular Signatures Associated with African Ancestry in Triple Negative Breast Cancer Using Quantified Genetic Ancestry Models in Admixed Race Populations. Cancers . [Internet]. 2020;12(5):1220. Disponible en: https://doi.org/10.3390/cancers12051220
- Jackisch C, Harbeck N, Huober J, Von Minckwitz G, Gerber B, Kreipe HH, et al. 14th St. Gallen International Breast Cancer Conference 2015: Evidence, Controversies, Consensus - Primary Therapy of Early Breast Cancer: Opinions Expressed by German Experts. Breast Care [Internet]. 2015;10:211–9. Disponible en: https://doi.org/10.1159/000433590
- Parise CA, Bauer KR, Caggiano V. Variation in breast cancer subtypes with age and race/ethnicity. Crit Rev Oncol Hematol [Internet]. 2010;76:44–52. Disponible en: https://doi.org/10.1016/j.critrevonc.2009.09.002
- Roelands J, Mall R, Almeer H, Thomas R, Mohamed MG, Bedri S, et al. Ancestry-associated transcriptomic profiles of breast cancer in patients of African, Arab, and European ancestry. npj Breast Cancer 2021 7:1 [Internet]. 2021;7:1–14. Disponible en: https://doi.org/10.1038/s41523-021-00215-x
- Serrano-Gómez SJ, Sanabria-Salas MC, Garay J, Baddoo MC, Hernández-Suarez G, Mejía JC, et al. Ancestry as a potential modifier of gene expression in breast tumors from Colombian women. Toland AE, editor. PLoS One [Internet]. 2017;12:e0183179. Disponible en: https://doi.org/10.1371/journal.pone.0183179
- Marker KM, Zavala VA, Vidaurre T, Lott PC, Vásquez JN, Casavilca-Zambrano S, et al. Human epidermal growth factor receptor 2–positive breast cancer is associated with Indigenous American ancestry in Latin American women. Cancer Res [Internet]. 2020;80:1893–901. Disponible en: https://doi.org/10.1158/0008-5472.CAN-19-3659
- Rey-Vargas L, Bejarano-Rivera LM, Mejia-Henao JC, Sua LF, Bastidas-Andrade JF, Ossa CA, et al. Association of genetic ancestry with HER2, GRB7 AND estrogen receptor expression among Colombian women with breast cancer. Front Oncol [Internet]. 2022;12:989761. Disponible en: https://doi.org/10.3389/fonc.2022.989761
- Alves da Quinta D, Rocha D, Yáñez C, Binato R, Soares-Lima SC, Huang X, et al. Genetic Ancestry, Intrinsic Tumor Subtypes, and Breast Cancer Survival in Latin American Women. Cancer research communications [Internet]. 2025;5:1070–81. Disponible en: https://doi.org/10.1158/2767-9764.CRC-25-0014
- da Costa Vieira RA, Sant’Anna D, Laus AC, Bacchi CE, Silva RJC, de Oliveira-Junior I, et al. Genetic Ancestry of 1127 Brazilian Breast Cancer Patients and Its Correlation With Molecular Subtype and Geographic Region. Clin Breast Cancer [Internet]. 2023;23:527–37. Disponible en: https://doi.org/10.1016/j.clbc.2023.04.001
- Rajagopal PS, Reid S, Fan R, Venton L, Weidner A, Roberson ML, et al. Population-specific patterns in assessing molecular subtypes of young black females with triple-negative breast cancer. NPJ Breast Cancer [Internet]. 2025;11:1–9. Disponible en: https://doi.org/10.1038/s41523-025-00731-0
- Thorn GJ, Gadaleta E, Dayem Ullah AZM, James LGE, Abdollahyan M, Barrow-McGee R, et al. The clinical and molecular landscape of breast cancer in women of African and South Asian ancestry. Nature Communications. [Internet]. 2025;16:4237. Disponible en: https://doi.org/10.1038/s41467-025-59144-z
- van Wilpe S, Tolmeijer SH, Koornstra RHT, de Vries IJM, Gerritsen WR, Ligtenberg M, et al. Homologous Recombination Repair Deficiency and Implications for Tumor Immunogenicity. Cancers 2021, Vol 13, Page 2249 [Internet]. 2021;13:2249. Disponible en: https://doi.org/10.3390/cancers13092249
- Paixão D, Torrezan GT, Santiago KM, Formiga MN, Ahuno ST, Dias-Neto E, et al. Characterization of genetic predisposition to molecular subtypes of breast cancer in Brazilian patients. Front Oncol [Internet]. 2022;12:976959. Disponible en: https://doi.org/10.3389/fonc.2022.976959
- Rujchanarong D, Spruill L, Sandusky GE, Park Y, Mehta AS, Drake RR, et al. Spatial N-glycomics of the normal breast microenvironment reveals fucosylated and high-mannose N-glycan signatures related to BI-RADS density and ancestry. Glycobiology [Internet]. 2024;34. Disponible en: https://doi.org/10.1093/glycob/cwae043
- Castañeda-González JP, Parra-Medina R, Riess JW, Gandara DR, Carvajal-Carmona LG. Genetic Ancestry and Lung Cancer in Latin American Patients: A Crucial Step for Understanding a Diverse Population. Clin Lung Cancer [Internet]. 2025;26:e342–52. Disponible en: https://doi.org/10.1016/j.cllc.2025.03.004
- Guo H, Li H, Zhu L, Feng J, Huang X, Baak JPA. “How Long Have I Got?” in Stage IV NSCLC Patients With at Least 3 Months Up to 10 Years Survival, Accuracy of Long-, Intermediate-, and Short-Term Survival Prediction Is Not Good Enough to Answer This Question. Front Oncol [Internet]. 2021;11. Disponible en: https://doi.org/10.3389/fonc.2021.761042
- Howlader N, Forjaz G, Mooradian MJ, Meza R, Kong CY, Cronin KA, et al. The Effect of Advances in Lung-Cancer Treatment on Population Mortality. New England Journal of Medicine [Internet]. 2020;383:640–9. Disponible en: https://doi.org/10.1056/NEJMoa1916623
- Campbell JD, Lathan C, Sholl L, Ducar M, Vega M, Sunkavalli A, et al. Comparison of Prevalence and Types of Mutations in Lung Cancers Among Black and White Populations. JAMA Oncol [Internet]. 2017;3:801–9. Disponible en: https://doi.org/10.1001/jamaoncol.2016.6108
- Arauz RF, Byun JS, Tandon M, Sinha S, Kuhn S, Taylor S, et al. Whole-Exome Profiling of NSCLC Among African Americans. Journal of Thoracic Oncology [Internet]. 2020;15:1880–92. Disponible en: https://doi.org/10.1016/j.jtho.2020.08.029
- Araujo LH, Timmers C, Bell EH, Shilo K, Lammers PE, Zhao W, et al. Genomic characterization of non-small-cell lung cancer in African Americans by targeted massively parallel sequencing. Journal of Clinical Oncology [Internet]. 2015;33(17):1966–73. Disponible en: https://doi.org/10.1200/JCO.2014.59.2444
- Chen J, Yang H, Teo ASM, Amer LB, Sherbaf FG, Tan CQ, et al. Genomic landscape of lung adenocarcinoma in East Asians. Nature Genetics [Internet]. 2020;52:177–86. Disponible en: https://doi.org/10.1038/s41588-019-0569-6
- Rhead B, Pouliot Y, Guinney J, Vega FMD La. Genetic Ancestry and Somatic Mutations in Lung Adenocarcinoma: Insights from Real-World Clinico-Genomic Data. medRxiv [Internet]. 2024;2024.04.24.24306316. Disponible en: https://doi.org/10.1101/2024.04.24.24306316
- Adib E, Nassar AH, Abou Alaiwi S, Groha S, Akl EW, Sholl LM, et al. Variation in targetable genomic alterations in non-small cell lung cancer by genetic ancestry, sex, smoking history, and histology. Genome Med [Internet]. 2022;14:1–10. Disponible en: https://doi.org/10.1186/s13073-022-01041-x
- Arrieta O, Cardona AF, Federico Bramuglia G, Gallo A, Campos-Parra AD, Serrano S, et al. Genotyping non-small cell lung cancer (NSCLC) in latin America. Journal of Thoracic Oncology [Internet]. 2011;6:1955–9. Disponible en: https://doi.org/10.1097/JTO.0b013e31822f655f
- Carrot-Zhang J, Soca-Chafre G, Patterson N, Thorner AR, Nag A, Watson J, et al. Genetic ancestry contributes to somatic mutations in lung cancers from admixed latin american populations. Cancer Discov [Internet]. 2021;11(3):591–8. Disponible en: https://doi.org/10.1158/2159-8290.CD-20-1165
- de Oliveira Cavagna R, Escremim de Paula F, Berardinelli GN, Bonatelli M, Santana I, Albino da Silva EC, et al. Molecular profile of driver genes in lung adenocarcinomas of Brazilian patients who have never smoked: implications for targeted therapies. Oncologist [Internet]. 2024;29:e1419–24. Disponible en: https://doi.org/10.1093/oncolo/oyae129
- Vieira IA, Andreis TF, Fernandes BV, Achatz MI, Macedo GS, Schramek D, et al. Prevalence of the Brazilian TP53 Founder c.1010G>A (p.Arg337His) in Lung Adenocarcinoma: Is Genotyping Warranted in All Brazilian Patients? Front Genet [Internet]. 2021;12:606537. Disponible en: https://doi.org/10.3389/fgene.2021.606537
- Leal LF, de Paula FE, De Marchi P, de Souza Viana L, Pinto GDJ, Carlos CD, et al. Mutational profile of Brazilian lung adenocarcinoma unveils association of EGFR mutations with high Asian ancestry and independent prognostic role of KRAS mutations. Sci Rep [Internet]. 2019;9:1–10. Disponible en: https://doi.org/10.1038/s41598-019-39965-x
- Climente-González H, Porta-Pardo E, Godzik A, Eyras E. The Functional Impact of Alternative Splicing in Cancer. Cell Rep [Internet]. 2017;20(9):2215–26. Disponible en: https://doi.org/10.1016/j.celrep.2017.08.012
- Deveaux AE, Allen TA, Al Abo M, Qin X, Zhang D, Patierno BM, et al. RNA splicing and aggregate gene expression differences in lung squamous cell carcinoma between patients of West African and European ancestry. Lung Cancer [Internet]. 2021;153:90–8. Disponible en: https://doi.org/10.1016/j.lungcan.2021.01.015
- Bae JM, Kim JH, Kang GH. Molecular subtypes of colorectal cancer and their clinicopathologic features, with an emphasis on the serrated neoplasia pathway. Arch Pathol Lab Med [Internet]. 2016;140:406–12. Disponible en: https://doi.org/10.5858/arpa.2015-0310-RA
- De Palma FDE, D’argenio V, Pol J, Kroemer G, Maiuri MC, Salvatore F. The molecular hallmarks of the serrated pathway in colorectal cancer. Cancers (Basel) [Internet]. 2019;11. Disponible en: https://doi.org/10.3390/cancers11071017
- Benedix F, Kube R, Meyer F, Schmidt U, Gastinger I, Lippert H. Comparison of 17,641 patients with right- and left-sided colon cancer: Differences in epidemiology, perioperative course, histology, and survival. Dis Colon Rectum [Internet]. 2010;53:57–64. Disponible en: https://doi.org/10.1007/DCR.0b013e3181c703a4
- Rhead B, Hein DM, Pouliot Y, Guinney J, De La Vega FM, Sanford NN. Association of genetic ancestry with molecular tumor profiles in colorectal cancer. Genome Med [Internet]. 2024;16:1–17. Disponible en: https://doi.org/10.1186/s13073-024-01373-w
- Myer PA, Lee JK, Madison RW, Pradhan K, Newberg JY, Isasi CR, et al. The Genomics of Colorectal Cancer in Populations with African and European Ancestry. Cancer Discov [Internet]. 2022;12:1282–93. Disponible en: https://doi.org/10.1158/2159-8290.CD-21-0813
- Hernandez-Suarez G, Sanabria MC, Serrano M, Herran OF, Perez J, Plata JL, et al. Genetic ancestry is associated with colorectal adenomas and adenocarcinomas in Latino populations. European Journal of Human Genetics [Internet]. 2014;22:1208–16. Disponible en: https://doi.org/10.1038/ejhg.2013.310
- Berardinelli GN, Durães R, Mafra da Costa A, Bragagnoli A, Antônio de Oliveira M, Pereira R, et al. Association of microsatellite instability (MSI) status with the 5-year outcome and genetic ancestry in a large Brazilian cohort of colorectal cancer. European Journal of Human Genetics [Internet]. 2022;30:824–32. Disponible en: https://doi.org/10.1038/s41431-022-01104-y
- Sanabria-Salas MC, Hernández-Suárez G, Umaña-Pérez A, Rawlik K, Tenesa A, Serrano-López ML, et al. IL1B-CGTC haplotype is associated with colorectal cancer in admixed individuals with increased African ancestry. Sci Rep [Internet]. 2017;7:1–11. Disponible en: https://doi.org/10.1038/srep41920
- Durães RO, Berardinelli GN, da Costa AM, Scapulatempo-Neto C, Pereira R, Oliveira MA, et al. Role of Genetic Ancestry in 1,002 Brazilian Colorectal Cancer Patients From Barretos Cancer Hospital. Front Oncol [Internet]. 2020;10:494547. Disponible en: https://doi.org/10.3389/fonc.2020.00145
- Matejcic M, Teer JK, Hoehn HJ, Diaz DB, Shankar K, Gong J, et al. Colorectal Tumors in Diverse Patient Populations Feature a Spectrum of Somatic Mutational Profiles. Cancer Res [Internet]. 2025;85:1928–44. Disponible en: https://doi.org/10.1158/0008-5472.CAN-24-0747
- dos Santos W, Sobanski T, de Carvalho AC, Evangelista AF, Matsushita M, Berardinelli GN, et al. Mutation profiling of cancer drivers in Brazilian colorectal cancer. Scientific Reports 2019 9:1 [Internet]. 2019;9:1–13. Disponible en: https://doi.org/10.1038/s41598-019-49611-1
- dos Santos W, dos Reis MB, Porto J, de Carvalho AC, Matsushita M, Oliveira G, et al. Somatic targeted mutation profiling of colorectal cancer precursor lesions. BMC Med Genomics [Internet]. 2022;15:1–11. Disponible en: https://doi.org/10.1186/s12920-022-01294-w
- Curran T, Sun Z, Gerry B, Findlay VJ, Wallace K, Li Z, et al. Differential immune signatures in the tumor microenvironment are associated with colon cancer racial disparities. Cancer Med [Internet]. 2021;10:1805–14. Disponible en: https://doi.org/10.1002/cam4.3753
- Wallace K, Lewin DN, Sun S, Spiceland CM, Rockey DC, Alekseyenko A V., et al. Tumor-infiltrating lymphocytes and colorectal cancer survival in Africa Namerican and Caucasian Patients. Cancer Epidemiology Biomarkers and Prevention [Internet]. 2018;27:755–61. Disponible en: https://doi.org/10.1158/1055-9965.EPI-17-0870
- Myer PA, Kim H, Blümel AM, Finnegan E, Kel A, Thompson T V., et al. Master Transcription Regulators and Transcription Factors Regulate Immune-Associated Differences Between Patients of African and European Ancestry With Colorectal Cancer. Gastro Hep Advances [Internet]. 2022;1:328–41. Disponible en: https://doi.org/10.1016/j.gastha.2022.01.004
- Jaratlerdsiri W, Jiang J, Gong T, Patrick SM, Willet C, Chew T, et al. African-specific molecular taxonomy of prostate cancer. Nature 2022 609:7927 [Internet]. 2022;609:552–9. Disponible en: https://doi.org/10.1038/s41586-022-05154-6
- Okobia MN, Zmuda JM, Ferrell RE, Patrick AL, Bunker CH. Chromosome 8q24 variants are associated with prostate cancer risk in a high risk population of African ancestry. Prostate [Internet]. 2011;71:1054–63. Disponible en: https://doi.org/10.1002/pros.21320
- Pal G, Di L, Orunmuyi A, Oluwabunmi Olapade-Olaopa E, Qiu W, Ogunwobi OO. Population differentiation at the PVT1 gene locus: Implications for prostate cancer. G3: Genes, Genomes, Genetics [Internet]. 2020;10:2257–64. Disponible en: https://doi.org/10.1534/g3.120.401291
- Mendes AA, Lu J, Kaur HB, Zheng SL, Xu J, Hicks J, et al. Association of B7-H3 expression with racial ancestry, immune cell density, and androgen receptor activation in prostate cancer. Cancer [Internet]. 2022;128:2269–80. Disponible en: https://doi.org/10.1002/cncr.34190
- Du Z, Hopp H, Ingles SA, Huff C, Sheng X, Weaver B, et al. A genome-wide association study of prostate cancer in Latinos. Int J Cancer [Internet]. 2020;146:1819–26. Disponible en: https://doi.org/10.1002/ijc.32525
- Al Abo M, Foo WC, Howard LE, McGue S, Lacroix B, Kephart J, et al. Genetic ancestry concordant RNA splicing in prostate cancer involves oncogenic genes and associates with recurrence. NPJ Precis Oncol [Internet]. 2025;9:1–16. Disponible en: https://doi.org/10.1038/s41698-025-00817-9
- Taylor J, Xiao W, Abdel-Wahab O. Diagnosis and classification of hematologic malignancies on the basis of genetics. Blood [Internet]. 2017;130:410–23. Disponible en: https://doi.org/10.1182/blood-2017-02-734541
- Ward E, DeSantis C, Robbins A, Kohler B, Jemal A. Childhood and adolescent cancer statistics, 2014. CA Cancer J Clin [Internet]. 2014;64:83–103. Disponible en: https://doi.org/10.3322/caac.21219
- Yang JJ, Cheng C, Devidas M, Cao X, Fan Y, Campana D, et al. Ancestry and Pharmacogenomics of Relapse in Acute Lymphoblastic Leukemia. Nat Genet [Internet]. 2011;43:237-41. Disponible en: https://doi.org/10.1038/ng.763
- Newman H, Lee SHR, Pölönen P, Shraim R, Li Y, Liu H, et al. Impact of Genetic Ancestry on Genomics and Survival Outcomes in T-cell Acute Lymphoblastic Leukemia. Blood Cancer Discov [Internet]. 2025;6(5):412-24 Disponible en: https://doi.org/10.1158/2643-3230.BCD-25-0049
- de Carvalho DC, Wanderley AV, dos Santos AMR, Moreira FC, de Sá RBA, Fernandes MR, et al. Characterization of pharmacogenetic markers related to Acute Lymphoblastic Leukemia toxicity in Amazonian native Americans population. Sci Rep [Internet]. 2020;10:1–9. Disponible en: https://doi.org/10.1038/s41598-020-67312-y
- Xu H, Yang W, Perez-Andreu V, Devidas M, Fan Y, Cheng C, et al. Novel susceptibility variants at 10p12.31-12.2 for childhood acute lymphoblastic leukemia in ethnically diverse populations. J Natl Cancer Inst [Internet]. 2013;105:733–42. Disponible en: https://doi.org/10.1093/jnci/djt042
- Landgren O, Graubard BI, Katzmann JA, Kyle RA, Ahmadizadeh I, Clark R, et al. Racial disparities in the prevalence of monoclonal gammopathies: A population-based study of 12 482 persons from the national health and nutritional examination survey. Leukemia [Internet]. 2014;28:1537–42. Disponible en: https://doi.org/10.1038/leu.2014.34
- Kazandjian D, Hill E, Hultcrantz M, Rustad EH, Yellapantula V, Akhlaghi T, et al. Molecular underpinnings of clinical disparity patterns in African American vs. Caucasian American multiple myeloma patients. Blood Cancer J [Internet]. 2019;9:1–8. Disponible en: https://doi.org/10.1038/s41408-019-0177-9
- Manojlovic Z, Christofferson A, Liang WS, Aldrich J, Washington M, Wong S, et al. Comprehensive molecular profiling of 718 Multiple Myelomas reveals significant differences in mutation frequencies between African and European descent cases. PLoS Genet [Internet]. 2017;13. Disponible en: https://doi.org/10.1371/journal.pgen.1007087
- Martins Rodrigues F, Jasielec J, Perpich M, Kim A, Moma L, Li Y, et al. Germline predisposition in multiple myeloma. iScience [Internet]. 2025;28. Disponible en: https://doi.org/10.1016/j.isci.2024.111620
- Mezghani N, Yao A, Vasilyeva D, Kaplan N, Shackelford A, Yoon A, et al. Molecular Subtypes of Head and Neck Cancer in Patients of African Ancestry. Clin Cancer Res [Internet]. 2023;29(5):910-20. Disponible en: https://doi.org/10.1158/1078-0432.CCR-22-2258
- Weigelt B, Marra A, Selenica P, Rios-Doria E, Momeni-Boroujeni A, Berger MF, et al. Molecular Characterization of Endometrial Carcinomas in Black and White Patients Reveals Disparate Drivers with Therapeutic Implications. Cancer Discov [Internet]. 2023;13:2356–69. Disponible en: https://doi.org/10.1158/2159-8290.CD-23-0546
- Totoki Y, Saito-Adachi M, Shiraishi Y, Komura D, Nakamura H, Suzuki A, et al. Multiancestry genomic and transcriptomic analysis of gastric cancer. Nat Genet [Internet]. 2023;55:581–94. Disponible en: https://doi.org/10.1038/s41588-023-01333-x