Molecular biology of prostate cancer
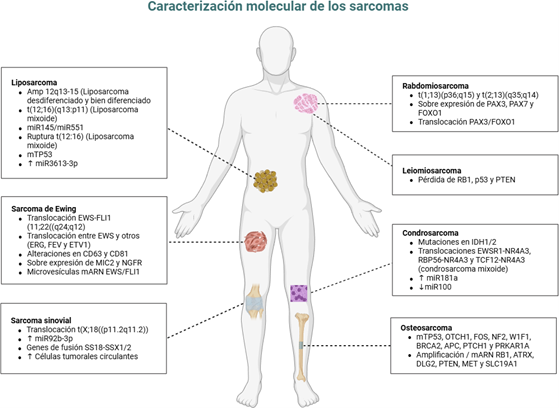
Perfilamiento molecular avanzado en sarcomas: claves para la práctica oncológica moderna
How to Cite
Download Citation

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Show authors biography
Introduction: prostate cancer (PCa) is the most common malignant neoplasm in men and a significant cause of cancer-related deaths. Its heterogeneity, ranging from indolent to aggressive forms, complicates risk stratification and therapeutic selection. However, advances in molecular biology have clarified the genomic and epigenetic alterations involved in tumorigenesis, progression, and resistance, thereby driving the development of precision medicine.
Methods: a narrative literature review was conducted on the key molecular pathways, clinically relevant genomic alterations, and emerging biomarker-based therapeutic strategies for localized and metastatic PCa.
Results: germline and somatic mutations in DNA repair genes, such as BRCA1, BRCA2, and ATM, are associated with increased susceptibility, aggressive phenotypes, and sensitivity to PARP inhibitors and platinum-based regimens. Loss of tumor suppressor genes (PTEN, TP53, and RB1) promotes genomic instability, castration resistance, and unfavorable prognosis. Genomic classifiers (Oncotype DX, Decipher) refine risk stratification and guide therapeutic intensification or de-escalation in localized disease, whereas reflex genetic testing and biomarker-driven clinical trials exemplify the clinical integration of molecular data. In advanced disease, therapies targeting androgen receptors, DNA repair mechanisms, and PSMA are transforming PCa management.
Conclusions: molecular characterization of PCa enables biomarker-guided interventions that optimize therapeutic decisions and clinical outcomes. The systematic incorporation of genomic profiling is essential for consolidating precision medicine in treatment strategies.
Article visits 0 | PDF visits 0
Downloads
- Pestana RC, Lopes David BB, Pires de Camargo V, Munhoz RR, Lopes de Mello CA, González Donna ML, et al. Challenges and opportunities for sarcoma care and research in Latin America: a position paper from the LACOG sarcoma group. Lancet Reg Health Am [Internet]. 2024;30:100671. Disponible en: https://doi.org/10.1016/j.lana.2023.100671
- Siegel RL, Miller KD, Wagle NS, Jemal A. Cancer statistics, 2023. CA Cancer J Clin [Internet]. 2023;73(1):17–48. Disponible en: https://doi.org/10.3322/caac.21763
- Borden EC, Baker LH, Bell RS, Bramwell V, Demetri GD, Eisenberg BL, et al. Soft tissue sarcomas of adults: state of the translational science. Clin Cancer Res [Internet]. 2003;9(6):1941–1956. PMID: 12796356
- Aponte-Monsalve J, Bolaños-Losada F. Avances y futuro del tratamiento de los sarcomas. Medicina (B Aires) [Internet]. 2024;46(2):451–471. Disponible en: https://doi.org/10.56050/01205498.2376
- Mitelman F. Recurrent chromosome aberrations in cancer. Mutat Res [Internet]. 2000;462(2–3):247–253. Disponible en: https://doi.org/10.1016/S1383-5742(00)00006-5
- Barr FG. Translocations, cancer and the puzzle of specificity. Nat Genet [Internet]. 1998;19(2):121–124. Disponible en: https://doi.org/10.1038/475
- Artandi SE, Chang S, Lee SL, Alson S, Gottlieb GJ, Chin L, DePinho RA. Telomere dysfunction promotes non-reciprocal translocations and epithelial cancers in mice. Nature [Internet]. 2000;406(6796):641–645. Disponible en: https://doi.org/10.1038/35020592
- Liu H, Wang X, Liu L, Yan B, Qiu F, Zhou B. Targeting liposarcoma: unveiling molecular pathways and therapeutic opportunities. Front Oncol [Internet]. 2024;14:1484027. Disponible en: https://doi.org/10.3389/fonc.2024.1484027
- Dwianingsih EK, Bawono RG, Saputri A, Malueka RG, Putro YAP, Anwar SL, et al. Histomorphological and molecular characteristics of liposarcoma (Review). Oncol Lett [Internet]. 2025;30(4):454. Disponible en: https://doi.org/10.3892/ol.2025.15200
- Segura Sánchez J, Pareja Megía MJ, García Escudero A, Vargas MT, González-Cámpora. R. Liposarcomas. Aspectos clínico-patológicos y moleculares. Rev Esp Patol [Internet]. 2006;39(3):135–148.11. Disponible en: https://doi.org/10.1016/S1699-8855(06)70028-9
- Jonczak E, Grossman J, Alessandrino F, Seldon Taswell C, Velez-Torres JM, Trent J. Liposarcoma: A Journey into a Rare Tumor's Epidemiology, Diagnosis, Pathophysiology, and Limitations of Current Therapies. Cancers (Basel). [Internet] 2024;16(22):3858. Disponible en: https://doi.org/10.3390/cancers16223858.
- Crago AM, Brennan MF. Principles in management of soft tissue sarcoma. Adv Surg [Internet]. 2015;49(1):107–122. Disponible en: https://doi.org/10.1016/j.yasu.2015.04.002
- Jamshidi F, Bashashati A, Shumansky K, Dickson B, Gokgoz N, Wunder JS, Andrulis IL, Lazar AJ, Shah SP, Huntsman DG, Nielsen TO. The genomic landscape of epithelioid sarcoma cell lines and tumours. J Pathol [Internet]. 2016;238(1):63–73. Disponible en: https://doi.org/10.1002/path.4636
- Lacuna K, Bose S, Ingham M, Schwartz G. Therapeutic advances in leiomyosarcoma. Front Oncol [Internet]. 2023;13:1149106. Disponible en: https://doi.org/10.3389/fonc.2023.1149106
- Guo X, Jo VY, Mills AM, Zhu SX, Lee CH, Espinosa I, et al. Clinically relevant molecular subtypes in leiomyosarcoma. Clin Cancer Res [Internet]. 2015;21(15):3501–3511. Disponible en: https://doi.org/10.1158/1078-0432.ccr-14-3141
- Falcao RM, Santana de Souza JES, Gonzalez-Molina J, Mathieson W, Carlson JW, Petta TB. Deep multi-omics integration approach reveals new molecular features of uterine leiomyosarcoma. Biochim Biophys Acta Mol Basis Dis [Internet]. 2025;1871(3): 167632. Disponible en: https://doi.org/10.1016/j.bbadis.2024.167632
- Haddox CL, Papke DJ Jr, Serrano C. Leiomyosarcoma therapeutic approaches and future directions. Hematol Oncol Clin North Am [Internet]. 2025;39(4):785–804. Disponible en: https://doi.org/10.1016/j.hoc.2025.04.007
- Grünewald TGP, Cidre-Aranaz F, Surdez D, Tomazou EM, de Álava E, Kovar H, et al. Ewing sarcoma. Nat Rev Dis Primers [Internet]. 2018;4(1):5. Disponible en: https://doi.org/10.1038/s41572-018-0003-x
- Kim SK, Park YK. Ewing sarcoma: a chronicle of molecular pathogenesis. Hum Pathol [Internet]. 2016;55:91–100. Disponible en: https://doi.org/10.1016/j.humpath.2016.05.008
- Subbiah V, Anderson P, Lazar AJ, Burdett E, Raymond K, Ludwig JA. Ewing’s sarcoma: standard and experimental treatment options. Curr Treat Options Oncol [Internet]. 2009;10(1–2):126–140. Disponible en: https://doi.org/10.1007/s11864-009-0104-6
- Kelleher FC, Thomas DM. Molecular pathogenesis and targeted therapeutics in Ewing sarcoma/primitive neuroectodermal tumours. Clin Sarcoma Res [Internet]. 2012;2(1):6. Disponible en: https://doi.org/10.1186/2045-3329-2-6
- Ozaki T, Schaefer KL, Wai D, Yokoyama R, Ahrens S, Diallo R, et al. Population-based genetic alterations in Ewing's tumors from Japanese and European Caucasian patients. Ann Oncol [Internet]. 2002;13(10):1656–1664. Disponible en: https://doi.org/10.1093/annonc/mdf218
- Parham DM, Barr FG. Classification of rhabdomyosarcoma and its molecular basis. Adv Anat Pathol [Internet]. 2013;20(6):387–397. Disponible en: https://doi.org/10.1097/PAP.0b013e3182a92d0d
- Dehner CA, Rudzinski ER, Davis JL. Rhabdomyosarcoma: updates on classification and the necessity of molecular testing beyond immunohistochemistry. Hum Pathol [Internet]. 2024;147:72–81. Disponible en: https://doi.org/10.1016/j.humpath.2023.12.004
- Franco ME, Aguabi SA, Medina AJ, Araujo ON, Gomez GM, Rabdomiosarcoma Infantil: Hallazgos clínicos – patológicos. Ciencia latina, revista multidisciplinar. Disponible en: https://doi.org/10.37811/cl_rcm.v7i1.5176
- Parham DM, Qualman SJ, Teot L, Barr FG, Morotti R, Sorensen PH, et al.; Soft Tissue Sarcoma Committee of the Children’s Oncology Group. Correlation between histology and PAX/FKHR fusion status in alveolar rhabdomyosarcoma: a report from the Children’s Oncology Group. Am J Surg Pathol [Internet]. 2007;31(6):895–901. Disponible en: https://doi.org/10.1097/01.pas.0000213436.99492.51
- Shenoy A, Alvarez E, Chi YY, Li M, Shern JF, Khan J, et al. The prognostic significance of anaplasia in childhood rhabdomyosarcoma: a report from the Children’s Oncology Group. Eur J Cancer [Internet]. 2021;143:127–133. Disponible en: https://doi.org/10.1016/j.ejca.2020.10.018
- Montiel JR. Mecanismos molecular de la diferenciación de células de rabdomiosarcoma [Internet]. Tesis doctoral. Universidad Nacional del Sur; 2018. Disponible en: https://repositoriodigital.uns.edu.ar/handle/123456789/7145
- Gallego S, Bernabeu D, Garrido-Pontnou M, Guillen G, Hindi N, Juan-Ribelles A, et al.; GEIS, SEHOP. GEIS-SEHOP clinical practice guidelines for the treatment of rhabdomyosarcoma. Clin Transl Oncol [Internet]. 2021;23(12):2460–2473. Disponible en: https://doi.org/10.1007/s12094-021-02 654-1
- Fiore M, Sambri A, Spinnato P, Zucchini R, Giannini C, Caldari E, et al. The biology of synovial sarcoma: state-of-the-art and future perspectives. Curr Treat Options Oncol [Internet]. 2021;22(12). Disponible en: https://doi.org/10.1007/s11864-021-00914-4
- Raquib AR, Hofvander J, Ta M, Nielsen TO. Expanding the use of an SS18-SSX antibody for molecular assays in synovial sarcoma. Appl Immunohistochem Mol Morphol [Internet]. 2022;30(8):531–539. Disponible en: https://doi.org/10.1097/PAI.0000000 000001049
- Lesovaya EA, Fetisov TI, Bokhyan BY, Maksimova VP, Kulikov EP, Belitsky GA, et al. Genetic and molecular heterogeneity of synovial sarcoma and associated challenges in therapy. Cells [Internet]. 2024;13(20):1695. Disponible en: https://doi.org/10.3390/cells13201695
- Wang AX, Jones KB, Nielsen TO. Molecular and epigenetic oncogenesis in synovial sarcoma: implications for cancer biology, diagnosis and treatment. Oncogene [Internet].2025;44(38):3527–3536. Disponible en: https://doi.org/10.1038/s41388-025-03 547-1
- Chen Y, Su Y, Cao X, Siavelis I, Leo IR, Zeng J, et al. Molecular profiling defines three subtypes of synovial sarcoma. Adv Sci (Weinh) [Internet]. 2024;11(41):e2404510. Disponible en: https://doi.org/10.1002/advs.202404510
- Floros KV, Fairchild CK Jr, Li J, Zhang K, Roberts JL, Kurupi R, et al. Targeting SUMOylation promotes cBAF complex stabilization and disruption of the SS18::SSX transcriptome in synovial sarcoma. Nat Commun [Internet]. 2025;16(1):9761. Disponible en: https://doi.org/10.1038/s41467-025-64665-8
- Chmiel P, Słowikowska A, Banaszek Ł, Szumera-Ciećkiewicz A, Szostakowski B, Spałek MJ, et al. Inflammatory myofibroblastic tumor from molecular diagnostics to current treatment. Oncol Res [Internet]. 2024;32(7):1141–1162. Disponible en: https://doi.org/10.32604/or.2024.050350
- Huang YL, Hsu CC, Huang DD, Yang JC, Wu SG. Identification of the molecular characterization and tumor microenvironment of thoracic inflammatory myofibroblastic tumors. J Formos Med Assoc [Internet]. 2025. Disponible en: https://doi.org/10.1016/j.jfma.2025.01.024
- Mahajan P, Casanova M, Ferrari A, Fordham A, Trahair T, Venkatramani R. Inflammatory myofibroblastic tumor: molecular landscape, targeted therapeutics, and remaining challenges. Curr Probl Cancer [Internet]. 2021;45(4):100768. Disponible en: [https://doi.org/10.1016/j.currproblcancer.2021.100768](https://doi.org/10.1016/j.currp roblcancer.2021.100768)
- Folpe AL, Deyrup AT. Alveolar soft-part sarcoma: a review and update. J Clin Pathol [Internet]. 2006;59(11):1127–1132. Disponible en: https://doi.org/10.1136/jcp.2005.031120
- Mello CA, Campos FAB, Santos TG, Silva MLG, Torrezan GT, Costa FD, et al. Desmoplastic small round cell tumor: a review of main molecular abnormalities and emerging therapy. Cancers (Basel) [Internet]. 2021;13(3):498. Disponible en: https://doi.org/10.3390/cancers13030498
- Henon C, Vibert J, Eychenne T, Gruel N, Colmet-Daage L, Ngo C, et al. Single-cell multiomics profiling reveals heterogeneous transcriptional programs and microenvironment in DSRCTs. Cell Rep Med [Internet]. 2024;5(6):101582. Disponible en: https://doi.org/10.1016/j.xcrm.2024.101582
- Czarnecka AM, Synoradzki K, Firlej W, Bartnik E, Sobczuk P, Fiedorowicz M, et al. Molecular biology of osteosarcoma. Cancers (Basel) [Internet]. 2020;12(8):2130. Disponible en: https://doi.org/10.3390/cancers12082130
- Hansen MF, Koufos A, Gallie BL, Phillips RA, Fodstad O, Brøgger A, et al. Osteosarcoma and retinoblastoma: a shared chromosomal mechanism revealing recessive predisposition. Proc Natl Acad Sci U S A [Internet]. 1985;82(18):6216–6220. Disponible en: https://doi.org/10.1073/pnas.82.18.6216
- Yu S, Yao X. Advances on immunotherapy for osteosarcoma. Mol Cancer [Internet]. 2024;23(1):192. Disponible en: https://doi.org/10.1186/s12943-024-02105-9
- Tlemsani C, Larousserie F, De Percin S, Audard V, Hadjadj D, Chen J, et al. Biology and management of high-grade chondrosarcoma: an update on targets and treatment options. Int J Mol Sci [Internet]. 2023;24(2):1361. Disponible en: https://doi.org/10.3390/ijms24021361
- Iacobescu GL, Corlatescu AD, Serban B, Spiridonica R, Costin HP, Cirstoiu C. Genetics and molecular pathogenesis of the chondrosarcoma: a review of the literature. Curr Issues Mol Biol [Internet]. 2024;46(11):12658–12671. Disponible en: https://doi.org/10.3390/cimb46110751
- Walker RL, Hornicek FJ, Duan Z. Advances in the molecular biology of chondrosarcoma for drug discovery and precision medicine. Cancers (Basel) [Internet]. 2025;17(16):2689. Disponible en: https://doi.org/10.3390/cancers17162689
- Ivanov S, Nano O, Hana C, Bonano-Rios A, Hussein A. Molecular targeting of the isocitrate dehydrogenase pathway and the implications for cancer therapy. Int J Mol Sci [Internet]. 2024;25(13):7337. Disponible en: https://doi.org/10.3390/ijms25137337
- De Vita A, Recine F, Mercatali L, Miserocchi G, Spadazzi C, Liverani C, et al. Primary culture of undifferentiated pleomorphic sarcoma: molecular characterization and response to anticancer agents. Int J Mol Sci [Internet]. 2017;18(12). Disponible en: https://doi.org/10.3390/ijms18122662
- Lu Y, Chen D, Wang B, Chai W, Yan M, Chen Y, et al. Single-cell landscape of undifferentiated pleomorphic sarcoma. Oncogene [Internet]. 2024;43(18):1353–1368. Disponible en: https://doi.org/10.1038/s41388-024-03001-8
- Serrano C, Romagosa C, Hernández-Losa J, Simonetti S, Valverde C, Moliné T, et al. RAS/MAPK pathway hyperactivation determines poor prognosis in undifferentiated pleomorphic sarcomas. Cancer [Internet]. 2016;122(1):99–107. Disponible en: https://doi.org/10.1002/cncr.29733
- Cazzato G, Piscazzi F, Filosa A, Colagrande A, Del Fiore P, Ambrogio F, et al. Clear cell sarcoma (CCS) of the soft tissue: an update narrative review with emphasis on the utility of PRAME in differential diagnosis. J Clin Med [Internet]. 2025;14(4):1233. Disponible en: https://doi.org/10.3390/jcm14041233
- Karner C, Anders I, Vejzovic D, Szkandera J, Scheipl S, Deutsch AJA, et al. Targeting epigenetic features in clear cell sarcomas based on patient-derived cell lines. J Transl Med [Internet] 2023;21(1):54. Disponible en: https://doi.org/10.1186/s12967-022-03843-4
- Rasmussen SV, Wozniak A, Lathara M, Goldenberg JM, Samudio BM, Bickford LR, et al. Functional genomics of human clear cell sarcoma: genomic, transcriptomic and chemical biology landscape for clear cell sarcoma. Br J Cancer [Internet]. 2023;128(10):1941–1954. Disponible en: https://doi.org/10.1038/s41416-023-02222-0
- Grünewald TGP, Postel-Vinay S, Nakayama RT, Berlow NE, Bolzicco A, Cerullo V, et al. Translational aspects of epithelioid sarcoma: current consensus. Clin Cancer Res [Internet]. 2024;30(6):1079–1092. Disponible en: https://doi.org/10.1158/1078-0432.ccr-23-2174
- Lualdi E, Modena P, Debiec-Rychter M, Pedeutour F, Teixeira MR, Facchinetti F, et al. Molecular cytogenetic characterization of proximal-type epithelioid sarcoma. Genes Chromosomes Cancer [Internet]. 2004;41(3):283–290. Disponible en: https://doi.org/10.1002/gcc.20086
- Haefliger S, Chervova O, Davies C, Nottley S, Hargreaves S, Sumathi VP, et al. Subclassification of epithelioid sarcoma with potential therapeutic impact. J Pathol [Internet]. 2023;260(4):368–375. Disponible en: https://doi.org/10.1002/path.6135
- Choi JH, Ro JY. The recent advances in molecular diagnosis of soft tissue tumors. Int J Mol Sci [Internet] 2023;24(6):5934. Disponible en: https://doi.org/10.3390/ijms24065934
- Cordier F, Ferdinande L, Hoorens A, Van de Vijver K, Van Dorpe J, Creytens D. Soft tissue and bone tumor diagnostics: harnessing the power of molecular techniques. Genes (Basel) [Internet]. 2023;14(12):2229. Disponible en: https://doi.org/10.3390/genes14122229