Single-cell analysis in cancer: clinical applications and roadmap for implementation
Análisis de célula única en cáncer: aplicaciones clínicas y hoja de ruta para su implementación
How to Cite
Download Citation

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Show authors biography
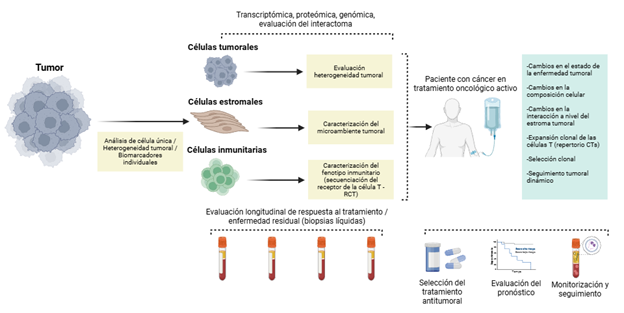
Introduction: single cell analysis comprises a set of technologies that enable the characterization of multiple omic layers including genomics, transcriptomics, epigenomics and proteomics at the level of individual cells. This approach provides a higher cellular resolution than bulk sequencing, which integrates heterogeneous signals into averaged molecular profiles. The incorporation of these platforms into oncology has reshaped the understanding of tumor heterogeneity, cellular plasticity and evolutionary processes underlying cancer development.
Methods: a narrative review of the literature was conducted. The bibliographic search was performed in PubMed/MEDLINE, EMBASE, Web of Science and Scopus. Inclusion criteria prioritized studies addressing clinical applications, particularly those related to immunotherapy, mechanisms of drug resistance, tumor heterogeneity and patient stratification.
Results: this review summarizes emerging single cell sequencing technologies and recent advances in cancer research derived from their application. The evidence synthesized includes findings related to malignant and immune cell landscapes, tumor heterogeneity, circulating tumor cells and biological mechanisms that shape tumor behavior.
Conclusion: collectively, available evidence indicates that unicellular approaches can provide clinically relevant value in diagnosis, therapeutic selection and prognostic assessment across multiple tumor types. Ongoing advances in this field are expected to further refine the biological characterization of tumors and to identify clinically actionable therapeutic targets.
Article visits 0 | PDF visits 0
Downloads
- 1. Hinohara K, Polyak K. Intratumoral Heterogeneity: More Than Just Mutations. Trends Cell Biol. 2019 Jul;29(7):569–79.
- 2. Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012 Jan;481(7381):306–13.
- 3. Merlo LMF, Pepper JW, Reid BJ, Maley CC. Cancer as an evolutionary and ecological process. Nat Rev Cancer. 2006 Dec;6(12):924–35.
- 4. Boxer E, Feigin N, Tschernichovsky R, Darnell NG, Greenwald AR, Hoefflin R, et al. Emerging clinical applications of single-cell RNA sequencing in oncology. Nat Rev Clin Oncol. 2025 May;22(5):315–26.
- 5. Tian J, Bai X, Quek C. Single-Cell Informatics for Tumor Microenvironment and Immunotherapy. Int J Mol Sci. 2024 Apr 19;25(8):4485.
- 6. Skinnider MA, Courtine G, Bloch J, Squair JW. A clinical road map for single-cell omics. Cell. 2025 Jul;188(14):3633–47.
- 7. Xu L, Saunders K, Huang SP, Knutsdottir H, Martinez-Algarin K, Terrazas I, et al. A comprehensive single-cell breast tumor atlas defines epithelial and immune heterogeneity and interactions predicting anti-PD-1 therapy response. Cell Rep Med. 2024 May;5(5):101511.
- 8. Chu Y, Dai E, Li Y, Han G, Pei G, Ingram DR, et al. Pan-cancer T cell atlas links a cellular stress response state to immunotherapy resistance. Nat Med. 2023 Jun;29(6):1550–62.
- 9. Aissa AF, Islam ABMMK, Ariss MM, Go CC, Rader AE, Conrardy RD, et al. Single-cell transcriptional changes associated with drug tolerance and response to combination therapies in cancer. Nat Commun. 2021 Mar 12;12(1):1628.
- 10. Griffiths JI, Cosgrove PA, Medina EF, Nath A, Chen J, Adler FR, et al. Cellular interactions within the immune microenvironment underpins resistance to cell cycle inhibition in breast cancers. Nat Commun. 2025 Mar 3;16(1):2132.
- 11. Madissoon E, Wilbrey-Clark A, Miragaia RJ, Saeb-Parsy K, Mahbubani KT, Georgakopoulos N, et al. scRNA-seq assessment of the human lung, spleen, and esophagus tissue stability after cold preservation. Genome Biol. 2020 Dec;21(1):1.
- 12. Denisenko E, Guo BB, Jones M, Hou R, De Kock L, Lassmann T, et al. Systematic assessment of tissue dissociation and storage biases in single-cell and single-nucleus RNA-seq workflows. Genome Biol. 2020 Dec;21(1):130.
- 13. Wang T, Roach MJ, Harvey K, Morlanes JE, Kiedik B, Al-Eryani G, et al. snPATHO-seq, a versatile FFPE single-nucleus RNA sequencing method to unlock pathology archives. Commun Biol. 2024 Oct 16;7(1):1340.
- 14. Fleming SJ, Chaffin MD, Arduini A, Akkad AD, Banks E, Marioni JC, et al. Unsupervised removal of systematic background noise from droplet-based single-cell experiments using CellBender. Nat Methods. 2023 Sep;20(9):1323–35.
- 15. Young MD, Behjati S. SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. GigaScience. 2020 Dec 26;9(12):giaa151.
- 16. Luecken MD, Büttner M, Chaichoompu K, Danese A, Interlandi M, Mueller MF, et al. Benchmarking atlas-level data integration in single-cell genomics. Nat Methods. 2022 Jan;19(1):41–50.
- 17. Horak P, Griffith M, Danos AM, Pitel BA, Madhavan S, Liu X, et al. Standards for the classification of pathogenicity of somatic variants in cancer (oncogenicity): Joint recommendations of Clinical Genome Resource (ClinGen), Cancer Genomics Consortium (CGC), and Variant Interpretation for Cancer Consortium (VICC). Genet Med Off J Am Coll Med Genet. 2022 May;24(5):986–98.
- 18. Westphalen CB, Boscolo Bielo L, Aftimos P, Beltran H, Benary M, Chakravarty D, et al. ESMO Precision Oncology Working Group recommendations on the structure and quality indicators for molecular tumour boards in clinical practice. Ann Oncol. 2025 Jun;36(6):614–25.
- 19. Klughammer J, Abravanel DL, Segerstolpe Å, Blosser TR, Goltsev Y, Cui Y, et al. A multi-modal single-cell and spatial expression map of metastatic breast cancer biopsies across clinicopathological features. Nat Med. 2024 Nov;30(11):3236–49.
- 20. Gondal MN, Cieslik M, Chinnaiyan AM. Integrated cancer cell-specific single-cell RNA-seq datasets of immune checkpoint blockade-treated patients. Sci Data. 2025 Jan 22;12(1):139.
- 21. Li W, Pan L, Hong W, Ginhoux F, Zhang X, Xiao C, et al. A single-cell pan-cancer analysis to show the variability of tumor-infiltrating myeloid cells in immune checkpoint blockade. Nat Commun. 2024 Jul 21;15(1):6142.
- 22. Sun X, Axelrod ML, Waks AG, Fu J, DiLullo M, Van Allen EM, et al. Dynamic single-cell systemic immune responses in immunotherapy-treated early-stage HR+ breast cancer patients. Npj Breast Cancer. 2025 Jul 3;11(1):65.
- 23. Oliveira MFD, Romero JP, Chung M, Williams SR, Gottscho AD, Gupta A, et al. High-definition spatial transcriptomic profiling of immune cell populations in colorectal cancer. Nat Genet. 2025 Jun;57(6):1512–23.
- 24. Fu S, Wang S, Si D, Li G, Gao Y, Liu Q. Benchmarking single-cell multi-modal data integrations [Internet]. 2025 [cited 2025 Sep 14]. Available from: http://biorxiv.org/lookup/doi/10.1101/2025.04.01.646578
- 25. Van Den Brink SC, Sage F, Vértesy Á, Spanjaard B, Peterson-Maduro J, Baron CS, et al. Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat Methods. 2017 Oct;14(10):935–6.
- 26. Cao K, Wei S, Ma T, Yang X, Wang Y, He X, et al. Integrating bulk, single-cell, and spatial transcriptomics to identify and functionally validate novel targets to enhance immunotherapy in NSCLC. Npj Precis Oncol. 2025 Apr 16;9(1):112.
- 27. Füllgrabe A, George N, Green M, Nejad P, Aronow B, Fexova SK, et al. Guidelines for reporting single-cell RNA-seq experiments. Nat Biotechnol. 2020 Dec;38(12):1384–6.
- 28. Kim GD, Lim C, Park J. A practical handbook on single-cell RNA sequencing data quality control and downstream analysis. Mol Cells. 2024 Sep;47(9):100103.
- 29. Walker CR, Li X, Chakravarthy M, Lounsbery-Scaife W, Choi YA, Singh R, et al. Private information leakage from single-cell count matrices. Cell. 2024 Nov;187(23):6537-6549.e10.