Tumor genomics: historical development and perspectives for the future
Genómica tumoral: construcción histórica y perspectivas para el futuro
How to Cite
Download Citation

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Show authors biography
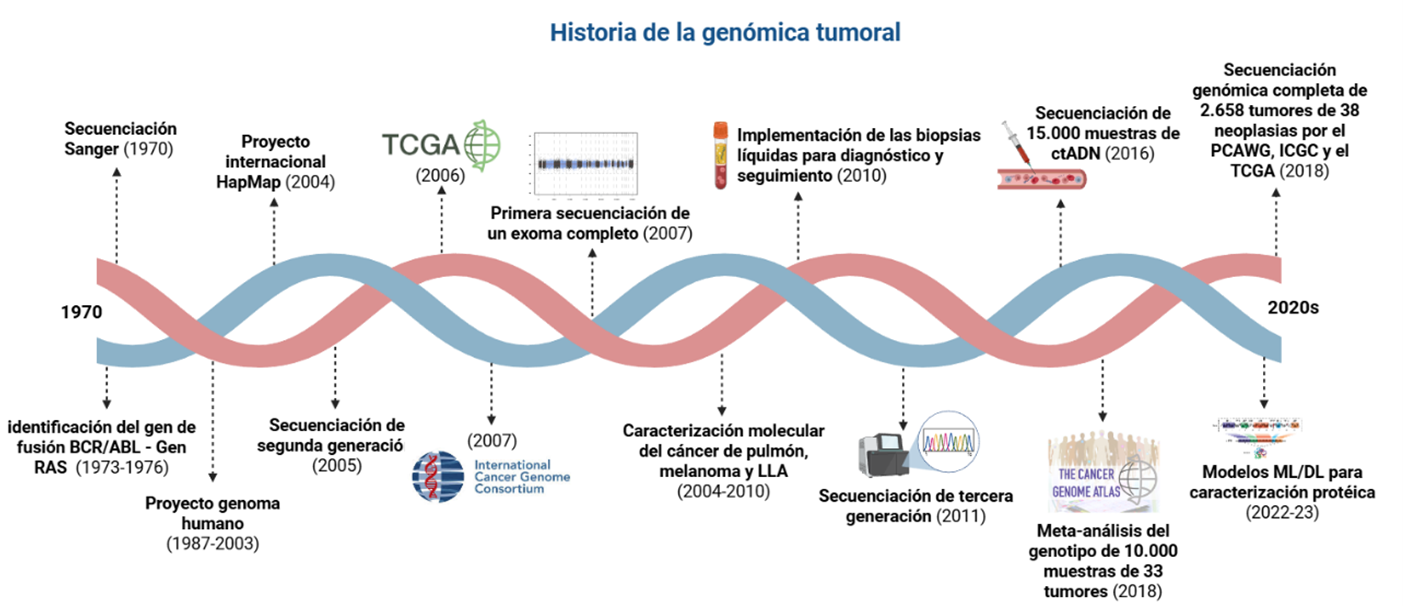
Introduction: from the discovery of the Philadelphia chromosome to the advent of next-generation sequencing (NGS), cancer genomics has transformed the molecular understanding of cancer. This article presents a narrative review with historical reconstruction, key milestones, major barriers, and future perspectives in tumor genomics.
Methods: a systematic search was conducted in three international databases; out of 137 records, 29 articles were selected for full review, along with various primary historical sources.
Results: this work examines technological evolution, clinical milestones, and the impact of genomics on oncology practice. In addition, some current challenges and future perspectives for its implementation were discussed.
Discussion: despite reviewing a time span of just over half a century, the research team acknowledges that these concepts are still constantly evolving and that overcoming current genomic barriers will likely be the next major milestone. Oncogenomics may be entering a golden era; its clinical implementation is increasingly becoming a reality and a standard.
Conclusion: in the era of massive data analysis, artificial intelligence, and robust global collaboration consortia, critically looking back at the past can also strengthen future steps. Cancer is now understood as a disease of the genome; however, it remains necessary to continue overcoming theoretical, scientific, implementation, accessibility, and cross-disciplinary obstacles.
Article visits 0 | PDF visits 0
Downloads
- Stafford N. Janet Rowley. BMJ [Internet]. 2014;348. Disponible en: https://doi.org/10.1136/bmj.g447
- Fonseca-Montaño MA, Blancas S, Herrera-Montalvo LA, Hidalgo-Miranda A. Cancer Genomics. Arch Med Res. [Internet]. 2022;53(8):723-31. Disponible en: https://doi.org/10.1016/j.arcmed.2022.11.011
- Yates LR, Campbell PJ. Evolution of the cancer genome. Nat Rev Genet. [Internet]. 2012;13(11):795-806. Disponible en: https://doi.org/10.1038/nrg3317
- Lee-Six H. Somatic evolution in human blood and colon. Doctoral tesis University of Cambridge. 2018. Disponible en: https://doi.org/10.17863/CAM.37059
- Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. [Internet]. 2009;458(7239):719-24. Disponible en: https://doi.org/10.1038/nature07943
- Nowell C. The minute chromosome (Ph1) in chronic granulocytic leukemia. Blut. [Internet]. 1962;8(2):65-6. Disponible en: https://doi.org/10.1007/bf01630378
- Rowley JD. A New Consistent Chromosomal Abnormality in Chronic Myelogenous Leukaemia identified by Quinacrine Fluorescence and Giemsa Staining. Nature. [Internet]. 1973;243(5405):290-3. Disponible en: https://doi.org/10.1038/243290a0
- Groffen J, Stephenson J, Heisterkamp N, Deklein A, Bartram C, Grosveld G. Philadelphia chromosomal breakpoints are clustered within a limited region, bcr, on chromosome 22. Cell. [Internet]. 1984;36(1):93-9. Disponible en: https://doi.org/10.1016/0092-8674(84)90077-1
- Spector DH, Smith K, Padgett T, McCombe P, Roulland-Dussoix D, Moscovici C, et al. Uninfected avian cells contain RNA related to the transforming gene of avian sarcoma viruses. Cell. [Internet]. 1978;13(2):371-9. Disponible en: https://doi.org/10.1016/0092-8674(78)90205-2
- Shampo MA, Kyle RA. J. Michael Bishop—Nobel Laureate in Medicine or Physiology. Mayo Clinic Proceedings. [Internet]. 2002;77(12):1312. Disponible en: https://doi.org/10.4065/77.12.1312
- Nowell PC. The Clonal Evolution of Tumor Cell Populations: Acquired genetic lability permits stepwise selection of variant sublines and underlies tumor progression. Science. [Internet]. 1976;194(4260):23-8. Disponible en: https://doi.org/10.1126/science.959840
- Rowley JD. Chromosome abnormalities in leukemia. J Clin Oncol. [Internet]. 1988;6(2):194-202. Disponible en: https://doi.org/10.1200/JCO.1988.6.2.194
- Ribeiro IP, Melo JB, Carreira IM. Cytogenetics and Cytogenomics Evaluation in Cancer. IJMS. [Internet]. 2019;20(19):4711. Disponible en: https://doi.org/10.3390/ijms20194711
- Mullis K, Faloona F, Scharf S, Saiki R, Horn G, Erlich H. Specific Enzymatic Amplification of DNA In Vitro: The Polymerase Chain Reaction. Cold Spring Harbor Symposia on Quantitative Biology. [Internet]. 1986;51(0):263-73. Disponible en: https://doi.org/10.1101/sqb.1986.051.01.032
- Slamon DJ, Leyland-Jones B, Shak S, Fuchs H, Paton V, Bajamonde A, et al. Use of Chemotherapy plus a Monoclonal Antibody against HER2 for Metastatic Breast Cancer That Overexpresses HER2. N Engl J Med. [Internet]. 2001;344(11):783-92. Disponible en: https://doi.org/10.1056/nejm200103153441101
- Sanger F, Nicklen S, Coulson AR. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. [Internet]. 1977;74(12):5463-7. Disponible en: https://doi.org/10.1073/pnas.74.12.5463
- International Human Genome Sequencing Consortium, Whitehead Institute for Biomedical Research, Center for Genome Research:, Lander ES, Linton LM, Birren B, Nusbaum C, et al. Initial sequencing and analysis of the human genome. Nature. [Internet]. 2001;409(6822):860-921. Disponible en: https://doi.org/10.1038/35079657
- Vogelstein B, Kinzler KW. The multistep nature of cancer. Trends in Genetics. [Internet]. 1993;9(4):138-41. Disponible en: https://doi.org/10.1016/0168-9525(93)90209-z
- Rubin MA, Demichelis F. The Genomics of Prostate Cancer: A Historic Perspective. Cold Spring Harb Perspect Med. [Internet]. 2019;9(3):a034942. Disponible en: https://doi.org/10.1101/cshperspect.a034942
- Casolino R, Beer PA, Chakravarty D, Davis MB, Malapelle U, Mazzarella L, et al. Interpreting and integrating genomic tests results in clinical cancer care: Overview and practical guidance. CA Cancer J Clin. [Internet]. 2024;74(3):264-85. Disponible en: https://doi.org/10.3322/caac.21825
- Brlek P, Bulić L, Bračić M, Projić P, Škaro V, Shah N, et al. Implementing Whole Genome Sequencing (WGS) in Clinical Practice: Advantages, Challenges, and Future Perspectives. Cells. [Internet]. 2024;13(6):504. Disponible en: https://doi.org/10.3390/cells13060504
- Heather JM, Chain B. The sequence of sequencers: The history of sequencing DNA. Genomics. [Internet]. 2016;107(1):1-8. Disponible en: https://doi.org/10.1016/j.ygeno.2015.11.003
- Torres-Narvaez ES, Mendivelso-González DF, Artunduaga-Alvarado JA, Ortega-Recalde O. Cancer genomics and bioinformatics in Latin American countries: applications, challenges, and perspectives. Front Oncol. [Internet]. 2025;15:1584178. Disponible en: https://doi.org/10.3389/fonc.2025.1584178
- Wang D, Liu B, Zhang Z. Accelerating the understanding of cancer biology through the lens of genomics. Cell. [Internet]. 2023;186(8):1755-71. Disponible en: https://doi.org/10.1016/j.cell.2023.02.015
- Berger MF, Mardis ER. The emerging clinical relevance of genomics in cancer medicine. Nat Rev Clin Oncol. [Internet]. 2018;15(6):353-65. Disponible en: https://doi.org/10.1038/s41571-018-0002-6
- De Las Salas B, Sánchez N, Gutiérrez L, Pérez S, Mercado V, Becerra P, et al. Investigación clínica en la era de la inmunoterapia y la genómica. Med. [Internet]. 2024;46(2):372-92. Disponible en: https://doi.org/10.56050/01205498.2371
- Sakamoto Y, Sereewattanawoot S, Suzuki A. A new era of long-read sequencing for cancer genomics. J Hum Genet. [Internet]. 2020;65(1):3-10. Disponible en: https://doi.org/10.1038/s10038-019-0658-5
- Valenti F, Falcone I, Ungania S, Desiderio F, Giacomini P, Bazzichetto C, et al. Precision Medicine and Melanoma: Multi-Omics Approaches to Monitoring the Immunotherapy Response. IJMS. [Internet]. 2021;22(8):3837. Disponible en: https://doi.org/10.3390/ijms22083837
- Testa U, Castelli G, Pelosi E. Breast Cancer: A Molecularly Heterogenous Disease Needing Subtype-Specific Treatments. Med Sci (Basel). [Internet]. 2020;8(1):18. Disponible en: https://doi.org/10.3390/medsci8010018
- Liskova A, Samec M, Koklesova L, Giordano FA, Kubatka P, Golubnitschaja O. Liquid Biopsy is Instrumental for 3PM Dimensional Solutions in Cancer Management. JCM. [Internet]. 2020;9(9):2749. Disponible en: https://doi.org/10.3390/jcm9092749
- Moldogazieva NT, Zavadskiy SP, Terentiev AA. Genomic Landscape of Liquid Biopsy for Hepatocellular Carcinoma Personalized Medicine. Cancer Genomics Proteomics. [Internet]. 2021;18(3 Suppl):369-83. Disponible en: https://doi.org/10.21873/cgp.20266
- Cammarata G, De Miguel-Perez D, Russo A, Peleg A, Dolo V, Rolfo C, et al. Emerging noncoding RNAs contained in extracellular vesicles: rising stars as biomarkers in lung cancer liquid biopsy. Ther Adv Med Oncol. [Internet]. 2022;14:17588359221131229. Disponible en: https://doi.org/10.1177/17588359221131229
- Black JRM, McGranahan N. Genetic and non-genetic clonal diversity in cancer evolution. Nat Rev Cancer. [Internet]. 2021;21(6):379-92. Disponible en: https://doi.org/10.1038/s41568-021-00336-2
- Mustachio LM, Roszik J. Single-Cell Sequencing: Current Applications in Precision Onco-Genomics and Cancer Therapeutics. Cancers. [Internet]. 2022;14(3):657. Disponible en: https://doi.org/10.3390/cancers14030657
- Bowes AL, Tarabichi M, Pillay N, Van Loo P. Leveraging single‐cell sequencing to unravel intratumour heterogeneity and tumour evolution in human cancers. The Journal of Pathology. [Internet]. 2022;257(4):466-78. Disponible en: https://doi.org/10.1002/path.5914
- Mulder NJ, Adebiyi E, Adebiyi M, Adeyemi S, Ahmed A, Ahmed R, et al. Development of Bioinformatics Infrastructure for Genomics Research. Glob Heart. [Internet]. 2017;12(2):91-98. Disponible en: https://globalheartjournal.com/article/10.1016/j.gheart.2017.01.005
- The ICGC/TCGA Pan-Cancer Analysis of Whole Genomes Consortium. Pan-cancer analysis of whole genomes. Nature. [Internet]. 2020;578(7793):82-93. Disponible en: https://doi.org/10.1038/s41586-020-1969-6
- Pan D, Jia D. Application of Single-Cell Multi-Omics in Dissecting Cancer Cell Plasticity and Tumor Heterogeneity. Front Mol Biosci. [Internet]. 2021;8:757024. Disponible en: https://doi.org/10.3389/fmolb.2021.757024
- Neinavaie F, Ibrahim-Hashim A, Kramer AM, Brown JS, Richards CL. The Genomic Processes of Biological Invasions: From Invasive Species to Cancer Metastases and Back Again. Front Ecol Evol. [Internet]. 2021;9:681100. Disponible en: https://doi.org/10.3389/fevo.2021.681100
- Williams ST, Wells G, Conroy S, Gagg H, Allen R, Rominiyi O, et al. Precision oncology using ex vivo technology: a step towards individualised cancer care? Expert Rev Mol Med. [Internet]. 2022;24:e39. Disponible en: https://doi.org/10.1017/erm.2022.32
- Cancer Genomics: The Road Ahead. Cell. [Internet]. 2013;155(1):9-10. Disponible en: https://linkinghub.elsevier.com/retrieve/pii/S0092867413011513
- McVeigh TP, Hughes LM, Miller N, Sheehan M, Keane M, Sweeney KJ, et al. The impact of Oncotype DX testing on breast cancer management and chemotherapy prescribing patterns in a tertiary referral centre. European Journal of Cancer. [Internet]. 2014;50(16):2763-70. Disponible en: https://doi.org/10.1016/j.ejca.2014.08.002
- Allemand C, Valerio AC, Calvo MF, Izbizky G, McLean I, Terrier F, et al. Impacto del Score de Recurrencia de 21 genes (Oncotype DX®) sobre la toma de decisión en tratamiento adyuvante: un estudio multicéntrico y colaborativo / Impact of the 21-gene Recurrence Score (Oncotype DX®) on decision-making in adjuvant treatment: a multicenter and collaborative study. Rev Argent Mastología [Internet]. 2024;42(154):13-27. Disponible en: https://doi.org/10.29289/2594539420210026
- Sparano JA, Gray RJ, Makower DF, Pritchard KI, Albain KS, Hayes DF, et al. Adjuvant Chemotherapy Guided by a 21-Gene Expression Assay in Breast Cancer. N Engl J Med. [Internet]. 2018;379(2):111-21. Disponible en: https://doi.org/10.1056/NEJMoa1804710
- Carugo A, Draetta GF. Academic Discovery of Anticancer Drugs: Historic and Future Perspectives. Annu Rev Cancer Biol. [Internet]. 2019;3(1):385-408. Disponible en: https://doi.org/10.1146/annurev-cancerbio-030518-055645
- Pich O, Bailey C, Watkins TBK, Zaccaria S, Jamal-Hanjani M, Swanton C. The translational challenges of precision oncology. Cancer Cell. [Internet]. 2022;40(5):458-78. Disponible en: https://doi.org/10.1016/j.ccell.2022.04.002
- O’Loughlin TA, Gilbert LA. Functional Genomics for Cancer Research: Applications In Vivo and In Vitro. Annu Rev Cancer Biol. [Internet]. 2019;3(1):345-63. Disponible en: https://doi.org/10.1146/annurev-cancerbio-030518-055742
- Kabadi A, McDonnell E, Frank CL, Drowley L. Applications of Functional Genomics for Drug Discovery. SLAS Discov. [Internet]. 2020;25(8):823-42. Disponible en: https://doi.org/10.1177/2472555220902092
- Li H, Yang Y, Hong W, Huang M, Wu M, Zhao X. Applications of genome editing technology in the targeted therapy of human diseases: mechanisms, advances and prospects. Sig Transduct Target Ther. [Internet]. 2020;5(1):1. Disponible en: https://doi.org/10.1038/s41392-019-0089-y
- Afolabi LO, Afolabi MO, Sani MM, Okunowo WO, Yan D, Chen L, et al. Exploiting the CRISPR‐Cas9 gene‐editing system for human cancers and immunotherapy. Clin & Trans Imm. [Internet]. 2021;10(6):e1286. Disponible en: https://doi.org/10.1002/cti2.1286
- Tran B, Dancey JE, Kamel-Reid S, McPherson JD, Bedard PL, Brown AMK, et al. Cancer Genomics: Technology, Discovery, and Translation. JCO. [Internet]. 2012;30(6):647-60. Disponible en: https://doi.org/10.1200/jco.2011.39.2316
- Vera J, Lai X, Baur A, Erdmann M, Gupta S, Guttà C, et al. Melanoma 2.0. Skin cancer as a paradigm for emerging diagnostic technologies, computational modelling and artificial intelligence. Briefings in Bioinformatics. [Internet]. 2022;23(6):bbac433. Disponible en: https://doi.org/10.1093/bib/bbac433
- Valent P, Orfao A, Kubicek S, Staber P, Haferlach T, Deininger M, et al. Precision Medicine in Hematology 2021: Definitions, Tools, Perspectives, and Open Questions. HemaSphere. [Internet]. 2021;5(3):e536. Disponible en: https://doi.org/10.1097/hs9.0000000000000536
- Ruíz-Patiño A. Futuro de la oncología personalizada y el diagnóstico molecular. Med. [Internet]. 2024;46(2):442-50. Disponible en: https://doi.org/10.56050/01205498.2375