New frontiers in precision oncology: transcriptomics and proteomics
Nuevas fronteras en la oncología de precisión: la transcriptómica y la proteómica
How to Cite
Download Citation

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Show authors biography
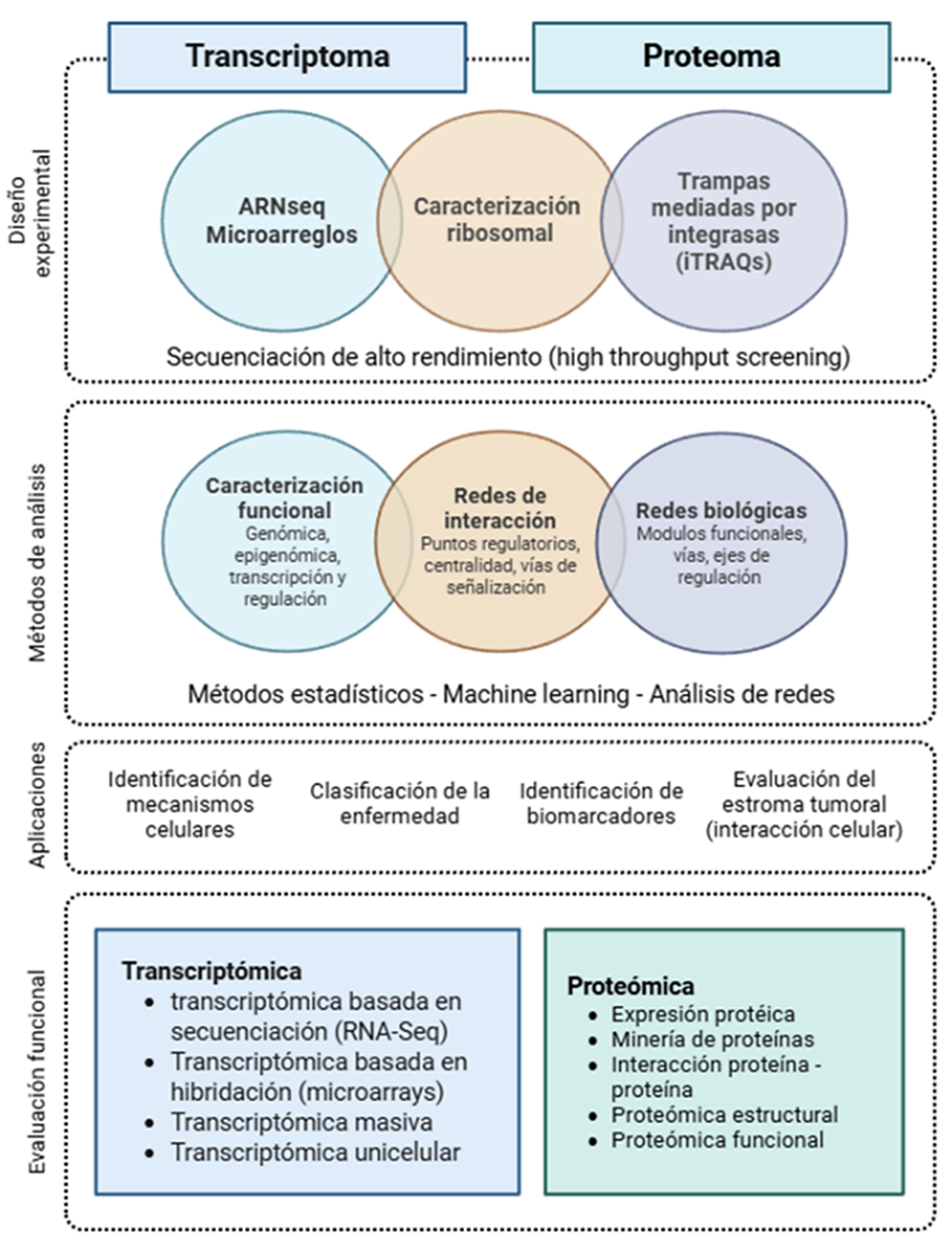
The integration of transcriptomics and proteomics in the study of cancer has significantly changed how we understand tumor biology, while also opening new possibilities for diagnosis and patient classification. Transcriptomics facilitates the identification of gene expression profiles, fusions, and the regulatory role of different non-coding RNAs in neoplastic progression; on the other hand, proteomics provides a more direct approach to cellular function, revealing active proteins and their post-translational modifications, crucial aspects in signaling mechanisms and treatment resistance. Although both disciplines have developed established clinical tools in precision oncology, limitations persist in reproducibility, standardization, and accessibility, particularly in resource-limited settings. Given this, integrative multi-omic approaches, supported by artificial intelligence and emerging technologies such as single-cell and spatial omics, are emerging as a promising way to reflect the biological complexity of cancer and advance toward dynamic diagnostics and personalized therapeutic strategies.
Article visits 0 | PDF visits 0
Downloads
- Kim HK, Kim T. Integrating Multi-Omics in Endometrial Cancer: From Molecular Insights to Clinical Applications. Cells. [Internet] 2025;8;14(17):1404-1014. Disponible en: https://doi.org/10.3390/cells14171404
- Sánchez-Bouza M, Sánchez-Frenes P, Ayala-Reina Z, Sánchez-Sánchez P, Santos-Solís M. Una mirada al cáncer desde la perspectiva molecular. Rev Finlay. [Internet] 2022;12(2):208–20. Disponible en: https://revfinlay.sld.cu/index.php/finlay/article/view/1027
- Zhu X, Zhao W, Zhou Z, Gu X. Unraveling the Drivers of Tumorigenesis in the Context of Evolution: Theoretical Models and Bioinformatics Tools. J Mol Evol. [internet] 2023;91(4):405-423. Disponible en: https://doi.org/10.1007/s00239-023-10117-0
- Merino JL, Guzmán G, Fernández-Cuadrado J. Ablación de la fibrilación auricular asistida por tomografía computarizada. Revista Española de Cardiología. [Internet] 2009;62(3):314-321. Disponible en: https://doi.org/10.1016/s0300-8932(09)70376-8
- Ubaid S, Kushwaha R, Kashif M, Singh V. Comprehensive analysis of oncogenic determinants across tumor types via multi-omics integration. Cancer Genetics. [internet] 2025;298-299:44-62. Disponible en https://doi.org/10.1016/j.cancergen.2025.08.010
- Martins Rodrigues F, Terekhanova NV, Imbach KJ, Porta-Pardo E, Ding L, et al. Precision proteogenomics reveals pan-cancer impact of germline variants. Cell. [Internet] 2025;188(9):2312-2335.e26. Disponible en: https://doi.org/10.1016/j.cell.2025.03.026
- Garg P, Krishna M, Kulkarni P, Horne D, Ravi Salgia, Singhal SS. Machine Learning Models for Predicting Gynecological Cancers: Advances, Challenges, and Future Directions. Cancers. [Internet] 2025;27(17):2790–9. Disponible en: https://doi.org/10.3390/cancers17172799
- Timms JF, Hale OJ, Cramer R. Advances in mass spectrometry-based cancer research and analysis: from cancer proteomics to clinical diagnostics. Expert Review of Proteomics. [Internet] 2016;13(6):593–607. Disponible en: https://doi.org/10.1080/14789450.2016.1182431
- Hajjo R, Sabbah DA, Bardaweel SK, Zhong HA. Targeting the EGFR/RAS/RAF signaling pathway in anticancer research: a recent update on inhibitor design and clinical trials (2020-2023). Expert opinion on therapeutic patents. [Internet] 2024;12:1–19. Disponible en: https://doi.org/10.1080/13543776.2024.2327307
- Fung JN, Pio R. Genetic variants in complement-related genes: potential implications for cancer risk and progression. Immunobiology. [Internet] 2025;230(4):153100. Disponible en: https://doi.org/10.1016/j.imbio.2025.153100
- Golub TR, Slonim DK, Tamayo P, Huard C, Gaasenbeek M, Mesirov JP, et al. Molecular classification of cancer: Class discovery and class prediction by gene expression monitoring. Science. [Internet] 1999;286(5439):531–7. Disponible en: https://doi.org/10.1126/science.286.5439.531
- Chen Z, He X. Application of third-generation sequencing in cancer research. Med Rev. [Internet] 2021;1(2):150-171. Disponible en: https://doi.org/10.1515/mr-2021-0013
- Wang Y, Mashock M, Tong Z, Mu X, Chen H, Zhou X, Zhang H, Zhao G, Liu B, Li X. Changing Technologies of RNA Sequencing and Their Applications in Clinical Oncology. Front Oncol. [Internet] 2020;10:447-456. Disponible en: https://doi.org/10.3389/fonc.2020.00447.
- Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. [Internet] 2009;10(1):57-63. Disponible en https://doi.org/10.1038/nrg2484.
- Byrne A, Beaudin AE, Olsen HE, Jain M, Cole C, Palmer T, et al. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat Commun. [Internet] 2017;8:16027-35. Disponible en: https://doi.org/10.1038/ncomms16027
- Schmidt F, Efferth T. Tumor Heterogeneity, Single-Cell Sequencing, and Drug Resistance. Pharmaceuticals. [Internet] 2016; 9(2):33-29. Disponible en: https://doi.org/10.3390/ph9020033
- Tang F, Barbacioru C, Wang Y, Nordman E, Lee C, Xu N, et al. mRNA-Seq whole-transcriptome analysis of a single cell. Nat Methods. [Internet] 2009;6(5):377–82. Disponible en: https://doi.org/10.1038/nmeth.1315
- Xiang L, Rao J, Yuan J, Xie T, Yan H. Single-Cell RNA-Sequencing: Opening New Horizons for Breast Cancer Research. International Journal of Molecular Sciences. [Internet] 2024; 25(17):9482-95. Disponible en: https://doi.org/10.3390/ijms25179482
- Xiang L, Rao J, Yuan J, Xie T, Yan H. Single-Cell RNA-Sequencing: Opening New Horizons for Breast Cancer Research. International Journal of Molecular Sciences. [Internet] 2024; 25(17):9482-90. Disponible en: https://doi.org/10.3390/ijms25179482
- Chen S, Zhou Z, Li Y, Du Y, Chen G. Application of single-cell sequencing to the research of tumor microenvironment. Front Immunol. [Internet] 2023;14:1285540. Disponible en: https://doi.org/10.3389/fimmu.2023.1285540
- Paik S, Shak S, Tang G, Kim C, Baker J, Cronin M, et al. A multigene assay to predict recurrence of tamoxifen-treated, node-negative breast cancer. N Engl J Med. [Internet] 2004;351(27):2817–26. Disponible en: https://doi.org/10.1056/nejmoa041588
- van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. [Internet] 2002;415(6871):530–6. Disponible en: https://doi.org/10.1038/415530a
- Huarte M. The emerging role of lncRNAs in cancer. Nat Med. [Internet] 2015;21:1253–61. Disponible en: https://doi.org/10.1038/nm.3981
- Chan YT, Zhang C, Wu J, Lu P, Xu L, Yuan H, Feng Y, Chen ZS, Wang N. Biomarkers for diagnosis and therapeutic options in hepatocellular carcinoma. Mol Cancer. [Internet] 2024;23:189. Disponible en https://doi.org/10.1186/s12943-024-02101-z
- Mohammadi E, Tahmoorespur M, Benfeitas R, Altay O, Javadmanesh A, et al. Improvement of the performance of anticancer peptides using a drug repositioning pipeline. Biotechnol J. [Internet] 2022;17(1):e2100417. Disponible en: https://doi.org/10.1002/biot.202100417
- Carrillo-Rodriguez P, Selheim F, Hernandez-Valladares M. Mass Spectrometry-Based Proteomics Workflows in Cancer Research: The Relevance of Choosing the Right Steps. Cancers (Basel). [Internet] 2023;15(2):555-60. Disponible en: https://doi.org/10.3390/cancers15020555
- Louati K, Kolsi F, Mellouli M, Louati H, Zribi R, Kallel R, et al. The Identification by Shotgun Proteomics with High-Resolution Tandem Mass-Spectrometry of Histone Isoforms' Hypermethylation Phenotype as a Hallmark Characteristic of Human-IDH-Mutant High-Grade Gliomas: Epigenetic Applications for Genotoxicity-Based Biomarkers and Cancer Therapy Targets. J Proteome Res. [Internet] 2025;24(9):4503-4525. Disponible en: https://doi.org/10.1021/acs.jproteome.5c00158.s001
- Mermelekas G, Vlahou A, Zoidakis J. SRM/MRM targeted proteomics as a tool for biomarker validation and absolute quantification in human urine. Expert Rev Mol Diagn. [Internet] 2015;15(11):1441-54. Disponible en: https://doi.org/10.1586/14737159.2015.1093937
- Ludwig C, Gillet L, Rosenberger G, Amon S, Collins BC, Aebersold R. Data-independent acquisition-based SWATH-MS for quantitative proteomics: a tutorial. Mol Syst Biol. [Internet] 2018;14(8):e8126. Disponible en: https://doi.org/10.15252/msb.20178126
- Naudot M, Le Ber J, Marcelo P. TMT-Based Quantitative Proteomics Analysis Reveals Differentially Expressed Proteins between Different Sources of hMSCs. International Journal of Molecular Sciences. [Internet] 2023; 24(17):13544. Disponible en: https://doi.org/10.3390/ijms241713544
- Mantini G, Pham TV, Piersma SR, Jimenez CR. Computational Analysis of Phosphoproteomics Data in Multi-Omics Cancer Studies. Proteomics. [Internet] 2021;21(3-4):e1900312. Disponible en: https://doi.org/10.1002/pmic.201900312
- Wu X, Xing X, Dowlut D, Zeng Y, Liu J, Liu X. Integrating phosphoproteomics into kinase-targeted cancer therapies in precision medicine. J Proteomics. [Internet] 2019;191:68-79. Disponible en: https://doi.org/10.1016/j.jprot.2018.03.033
- Aebersold R, Mann M. Mass-spectrometric exploration of proteome structure and function. Nature. [Internet] 2016;537(7620):347–55. Disponible en: https://doi.org/10.1038/nature19949
- Grzeski, M., Jensen, P. M., Hempel, B.-F., Thiele, H., Lellmann, J., Schallenberg, S., Budach, V., Keilholz, U., Tinhofer, I., & Klein, O. Integrating MALDI-MSI-Based Spatial Proteomics and Machine Learning to Predict Chemoradiotherapy Outcomes in Head and Neck Cancer. Int. J. Mol. Sci. [Internet] 2025; 26(18): 9084. Disponible en: https://doi.org/10.3390/ijms26189084
- Wu F, Yang J, Liu J, Wang Y, Mu J, Zeng Q, Deng S, Zhou H. Signaling pathways in cancer-associated fibroblasts and targeted therapy for cancer. Signal Transduct Target Ther. [Internet] 2021;6(1):218. Disponible en: https://doi.org/10.1038/s41392-021-00641-0
- Calderón-Rodríguez, Sandra Isabel, & Umaña-Pérez, Adriana. Estudio proteómico 2DE-DIGE en plasma sanguíneo de pacientes en etapa infantil con leucemia linfoblástica aguda. Revista Colombiana de Química. [Internet] 2019;48(1):5-15. Disponible en: https://doi.org/10.15446/rev.colomb.quim.v48n1.75170
- Reyes Tejera L, Fernández González OL, Alvarez Hernández JC. Biomarcadores salivales y su utilidad en la detección precoz del cáncer bucal. Rev. cuban. med. mil. [Internet] 2024;53(2): e024026095. Disponible en: http://scielo.sld.cu/scielo.php?script=sci_arttext&pid=S0138-65572024000200046&lng=es.
- Taguchi F, Solomon B, Gregorc V, Roder H, Gray R, Kasahara K, et al. Mass spectrometry to classify non–small-cell lung cancer patients for clinical outcome after treatment with epidermal growth factor receptor tyrosine kinase inhibitors: a multicohort cross-institutional study. J Natl Cancer Inst. [Internet] 2007;99(11):838–46. Disponible en: https://doi.org/10.1093/jnci/djk195
- Ueland FR, Desimone CP, Seamon LG, Miller RA, Goodrich S, Podzielinski I, et al. Effectiveness of a multivariate index assay in the preoperative assessment of ovarian tumors. Obstet Gynecol. [Internet] 2011;117(6):1289–97. Disponible en: https://doi.org/10.1097/aog.0b013e31821b5118
- Zhang H, Freitas D, Kim HS, Fabijanic K, Li Z, Chen H, et al. Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation. Nat Cell Biol. [Internet] 2018;20(3):332–43. Disponible en https://doi.org/10.1038/s41556-018-0040-4
- Ponomarenko EA, Krasnov GS, Kiseleva OI, Kryukova PA, Arzumanian VA, Dolgalev GV, Ilgisonis EV, Lisitsa AV, Poverennaya EV. Workability of mRNA Sequencing for Predicting Protein Abundance. Genes (Basel). [Internet] 2023;14(11):2065-72. Disponible en: https://doi.org/10.3390/genes14112065
- Zhang B, Wang J, Wang X, Zhu J, Liu Q, Shi Z, et al. Proteogenomic characterization of human colon and rectal cancer. Nature. [Internet] 2014;513:382–7. Disponible en: https://doi.org/10.1038/nature13438
- Zhang C, Li N, Zhang P, Jiang Z, Cheng Y, Li H, Pang Z. Advancing precision and personalized breast cancer treatment through multi-omics technologies. Am J Cancer Res. [Internet] 2024;14(12):5614-5627. Disponible en: https://doi.org/10.62347/mwnz5609
- Kerr K, McAneney H, Smyth LJ, Bailie C, McKee S, McKnight AJ. A scoping review and proposed workflow for multi-omic rare disease research. Orphanet J Rare Dis. [Internet] 2020;15(1):107. Disponible en: https://doi.org/10.1186/s13023-020-01376-x
- Huang S, Chaudhary K, Garmire LX. More is better: Recent progress in multi-omics data integration methods. Front Genet. [Internet] 2017;8:84. Disponible en: https://doi.org/10.3389/fgene.2017.00084
- Mertins P, Mani DR, Ruggles KV, Gillette MA, Clauser KR, Wang P, et al. Proteogenomics connects somatic mutations to signalling in breast cancer. Nature. [Internet] 2016;534:55–62. Disponible en: https://doi.org/10.1038/nature18003
- Ellis MJ, Gillette M, Carr SA, Paulovich AG, Smith RD, Rodland KK, et al. Connecting genomic alterations to cancer biology with proteomics: The NCI Clinical Proteomic Tumor Analysis Consortium. Cancer Discov. [Internet] 2013;3(10):1108–12. Disponible en: https://doi.org/10.1158/2159-8290.cd-13-0219
- Redig AJ, Jänne PA. Basket trials and the evolution of clinical trial design in an era of genomic medicine. J Clin Oncol. [Internet] 2015;33(9):975–7. Disponible en: https://doi.org/10.1200/jco.2014.59.8433
- Wang LB, Karpova A, Gritsenko MA, Kyle JE, Cao S, Li Y, et al. Proteogenomic and metabolomic characterization of human glioblastoma. Cancer Cell. [Internet] 2021;39(4):509–528.e20. Disponible en: https://doi.org/10.1016/j.ccell.2021.01.006
- Zhang H, Liu T, Zhang Z, Payne SH, Zhang B, McDermott JE, et al. Integrated proteogenomic characterization of human high-grade serous ovarian cancer. Cell. [Internet] 2025;188(24):7016. Disponible en: https://doi.org/10.1016/j.cell.2025.10.043
- Manuilova I, Bossenz J, Weise A, Boehm D, Döbel M, Werle S, et al. Uncovering the Understanding of the Concept of Patient Similarity in Cancer Research and Treatment: Scoping Review. J Med Internet Res. [Internet] 2025;27:e71906. Disponible en: https://doi.org/10.2196/71906
- Zou J, Wang E. Cancer Biomarker discovery for precision medicine: new progress. Curr Med Chem. [Internet] 2019;26(42):7655-7671. Disponible en: https://doi.org/10.2174/0929867325666180718164712
- Tirosh I, Izar B, Prakadan SM, Wadsworth MH, Treacy D, Trombetta JJ, et al. Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-seq. Science. [Internet] 2016;352:189–96. Disponible en: https://doi.org/10.1126/science.aad0501
- Marx V. Method of the year: spatially resolved transcriptomics. Nat Methods. [Internet] 2021;18:9–14. Disponible en: https://doi.org/10.1038/s41592-020-01033-y
- Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI. Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J. [Internet] 2015;13:8–17. Disponible en: https://doi.org/10.1016/j.csbj.2014.11.005
- Crowley E, Di Nicolantonio F, Loupakis F, Bardelli A. Liquid biopsy: monitoring cancer-genetics in the blood. Nat Rev Clin Oncol. [Internet] 2013;10(8):472–84. Disponible en: https://doi.org/10.1038/nrclinonc.2013.110
- Adesina A, Chumba D, Nelson AM, Orem J, Roberts DJ, Wabinga H, et al. Improvement of pathology in sub-Saharan Africa. Lancet Oncol. [Internet] 2013;14(4):e152–7. Disponible en: https://doi.org/10.1016/S1470-2045(12)70598-3