Bioinformatics in cancer
Bioinformática en cáncer
How to Cite
Download Citation

This work is licensed under a Creative Commons Attribution-NonCommercial-NoDerivatives 4.0 International License.
Show authors biography
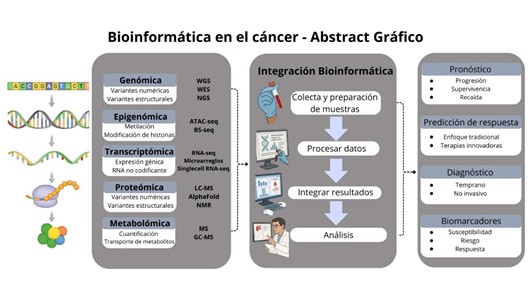
Introduction: Bioinformatics in cancer has become a powerful tool for the detection and monitoring of molecular variants associated with the disease. Currently, large amounts of omics data are handled to develop and apply new tools that allow efficient data analysis. The aim of this review was to describe the context in which new bioinformatics technologies have emerged and how these advances are contributing to cancer research.
Methods: A search was conducted in PubMed, Scopus, Google Scholar, and ScienceDirect. Information was presented based on a set of articles available in English and Spanish related to data analysis (n=66). In addition, the main databases and bioinformatics platforms for cancer research are presented.
Results: The reviewed studies show that bioinformatics acts as an integrative axis in tumor biology research, facilitating the identification of biomarkers, molecular classification of tumors, and patient stratification, while advances in artificial intelligence are revolutionizing data analysis.
Discussion: Bioinformatics tools have enabled a deeper understanding of the molecular mechanisms of cancer, supporting the development of strategies with potential clinical application; likewise, this increase in information requires stricter quality control.
Conclusion: Bioinformatics is an expanding tool, necessary in cancer research, that can facilitate appropriate guidance in diagnosis and treatment, impacting the natural history of the disease.
Article visits 0 | PDF visits 0
Downloads
- Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, Bray F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J Clin. [Internet]. 2021;71(3):209–49. Disponible en: https://doi.org/10.3322/caac.21660
- Topol EJ. High-performance medicine: the convergence of human and artificial intelligence. Nat. Med. [Internet]. 2019;25:44-56. Disponible en: https://doi.org/10.1038/s41591-018-0300-7
- Bode AM, Dong Z. Recent advances in precision oncology research. NPJ Precis Oncol [Internet]. 2018;2:11. Disponible en: https://doi.org/10.1038/s41698-018-0055-0
- Wang RC, Wang Z. Precision Medicine: Disease Subtyping and Tailored Treatment. Cancers [Internet]. 2023;15(15):3837. Disponible en: https://doi.org/10.3390/cancers15153837
- Molla G, Bitew M. Revolutionizing Personalized Medicine: Synergy with Multi-Omics Data Generation, Main Hurdles, and Future Perspectives. Biomedicines [Internet]. 2024;12(12):2750. Disponible en: https://doi.org/10.3390/biomedicines12122750
- Levy SE, Boone BE. Next-Generation sequencing Strategies. Cold Spring Harb Perspect Med [Internet]. 2018;9(7):a025791. Disponible en: https://doi.org/10.1101/cshperspect.a025791
- Satam H, Joshi K, Mangrolia U, Waghoo S, Zaidi G, Rawool S, et al. Next-Generation Sequencing Technology: Current Trends and Advancements. Biology [Internet]. 2023;12(7):997. Disponible en: https://doi.org/10.3390/biology12070997
- The International Cancer Genome Consortium. International network of cancer genome projects. Nature [Internet]. 2010;464(7291):993-8. Disponible en: https://doi.org/10.1038/nature08987
- International Human Genome Sequencing Consortium. . Initial sequencing and analysis of the human genome. Nature [Internet]. 2001;409:860-921. Disponible en: https://doi.org/10.1038/35087627
- Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, et al. The Sequence of the Human Genome. Science [Internet]. 2001;291(5507):1304-51. Disponible en: https://doi.org/10.1016/s0002-9394(01)01077-7
- Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz LA, Kinzler KW. Cancer genome landscapes. Science [Internet]. 2013;339(6127):1546-58. Disponible en: https://doi.org/10.1126/science.1235122
- Wood LD, Parsons DW, Jones S, Lin J, Sjöblom T, Leary RJ, et al. The Genomic Landscapes of Human Breast and Colorectal Cancers. Science [Internet]. 2007;318(5853):1108–13. Disponible en: http://dx.doi.org/10.1126/science.1145720
- Parsons DW, Jones S, Zhang X, Lin JCH, Leary RJ, Angenendt P, et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science [Internet]. 2008;321(5897):1807-12. Disponible en: https://doi.org/10.1126/science.1164382
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res [Internet]. 2010;20(9):1297-303. Disponible en: https://doi.org/10.1101/gr.107524.110
- Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, et al. Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature [Internet]. 2013;499(7457):214-8. Disponible en: https://doi.org/10.1038/nature12213
- Bailey MH, Tokheim C, Porta-Pardo E, Sengupta S, Bertrand D, Weerasinghe A, et al. Comprehensive Characterization of Cancer Driver Genes and Mutations. Cell [Internet]. 2018;173(2):371-385.e18. Disponible en: https://doi.org/10.1016/j.cell.2018.02.060
- Aaltonen LA, Abascal F, Abeshouse A, Aburatani H, Adams DJ, et al. Pan-cancer analysis of whole genomes. Nature [Internet]. 2020;578(7793):82-93. Disponible en: https://doi.org/10.1038/s41586-020-1969-6
- Tomczak K, Czerwińska P, Wiznerowicz M. Review The Cancer Genome Atlas (TCGA): an immeasurable source of knowledge. Contemp Oncol [Internet]. 2015;1A:68-77. Disponible en: https://doi.org/10.5114/wo.2014.47136
- Cosenza MR, Rodriguez-Martin B, Korbel JO. Structural Variation in Cancer: Role, Prevalence, and Mechanisms. Annu Rev Genom Hum Genet [Internet]. 2022;23(1):123-52. Disponible en: https://doi.org/10.1146/annurev-genom-120121-101149
- Regev A, Teichmann SA, Lander ES, Amit I, Benoist C, Birney E, et al. The Human Cell Atlas. eLife [Internet]. 2017;6. Disponible en: https://doi.org/10.7554/elife.27041
- Le J, Dian Y, Zhao D, Guo Z, Luo Z, Chen X, et al. Single-cell multi-omics in cancer immunotherapy: from tumor heterogeneity to personalized precision treatment. Mol Cancer [Internet]. 2025;24(1). Disponible en: https://doi.org/10.1186/s12943-025-02426-3
- Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA, et al. The cBio Cancer Genomics Portal: An Open Platform for Exploring Multidimensional Cancer Genomics Data. Cancer Discov [Internet]. 2012;2(5):401-4. Disponible en: https://doi.org/10.1158/2159-8290.cd-12-0095
- Leinonen R, Akhtar R, Birney E, Bower L, Cerdeno-Tarraga A, Cheng Y, et al. The European Nucleotide Archive. Nucleic Acids Res [Internet]. 2010;39:D28-31. Disponible en: https://doi.org/10.1093/nar/gkq967
- Leinonen R, Sugawara H, Shumway M. The sequence read archive. Nucleic Acids Res [Internet]. 2010;39:D19-21. Disponible en: https://doi.org/10.1093/nar/gkq1019
- Dyer SC, Austine-Orimoloye O, Azov AG, Barba M, Barnes I, Barrera-Enriquez VP, et al. Ensembl 2025. Nucleic Acids Res [Internet]. 2024;53(D1):D948-57. Disponible en: https://doi.org/10.1093/nar/gkae1071
- Perez G, Barber GP, Benet-Pages A, Casper J, Clawson H, Diekhans M, et al. The UCSC Genome Browser database: 2025 update. Nucleic Acids Res [Internet]. 2024;53(D1):D1243-9. Disponible en: https://doi.org/10.1093/nar/gkae974
- O’Leary NA, Cox E, Holmes JB, Anderson WR, Falk R, Hem V, et al. Exploring and retrieving sequence and metadata for species across the tree of life with NCBI Datasets. Sci. Data [Internet]. 2024;11(1):732. Disponible en: https://doi.org/10.1038/s41597-024-03571-y
- Forbes SA, Beare D, Boutselakis H, Bamford S, Bindal N, Tate J, et al. COSMIC: somatic cancer genetics at high-resolution. Nucleic Acids Res [Internet]. 2016;45(D1):D777-83. Disponible en: https://doi.org/10.1093/nar/gkw1121
- Landrum MJ, Lee JM, Riley GR, Jang W, Rubinstein WS, Church DM, Maglott DR. ClinVar: public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res [Internet]. 2014;42(1):D980-5. Disponible en: https://doi.org/10.1093/nar/gkt1113
- Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, et al. NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res [Internet]. 2013;41:D991-5. Disponible en: https://doi.org/10.1093/nar/gks1193
- Milacic M, Beavers D, Conley P, Gong C, Gillespie M, et al. The Reactome Pathway Knowledgebase 2024. Nucleic Acids Res [Internet]. 2024. Disponible en: https://doi.org/10.1093/nar/gkad1025
- Hoadley KA, Yau C, Hinoue T, Wolf DM, Lazar AJ, Drill E, et al. Cell-of-Origin Patterns Dominate the Molecular Classification of 10,000 Tumors from 33 Types of Cancer. Cell [Internet]. 2018;173(2):291-304.e6. Disponible en: https://doi.org/10.1016/j.cell.2018.03.022
- Hasin Y, Seldin M, Lusis A. Multi-omics approaches to disease. Genome Biol [Internet]. 2017;18(1). Disponible en: https://doi.org/10.1186/s13059-017-1215-1
- Kather JN, Heij LR, Grabsch HI, Loeffler C, Echle A, Muti HS, et al. Pan-cancer image-based detection of clinically actionable genetic alterations. Nat Cancer [Internet]. 2020;1(8):789-99. Disponible en: https://doi.org/10.1038/s43018-020-0087-6
- Nair AS. Computational Biology & Bioinformatics: A Gentle Overview. Communications of the Computer Society of India [Internet]. 2007;1–12. Disponible en: https://www.researchgate.net/publication/231337374_Computational_Biology_Bioinformatics_A_Gentle_Overview
- Bateman A, Martin MJ, Orchard S, Magrane M, Ahmad S, Alpi E, et al. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res [Internet]. 2022;51(D1):D523-31. Disponible en: https://doi.org/10.1093/nar/gkac1052
- Bateman A, Martin MJ, Orchard S, Magrane M, Ahmad S, Alpi E, et al. UniProt: the Universal Protein Knowledgebase in 2023. Nucleic Acids Res [Internet]. 2022;51(D1):D523-31. Disponible en: https://doi.org/10.1093/nar/gkac1052
- Van Der Auwera GA, Carneiro MO, Hartl C, Poplin R, Del Angel G, Levy‐Moonshine A, et al. From FastQ Data to High‐Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Curr Protoc Bioinformatics [Internet]. 2013;43(1). Disponible en: https://doi.org/10.1002/0471250953.bi1110s43
- Gonzalez-Perez A, Perez-Llamas C, Deu-Pons J, Tamborero D, Schroeder MP, Jene-Sanz A, et al. IntOGen-mutations identifies cancer drivers across tumor types. Nat Methods [Internet]. 2013;10(11):1081-2. Disponible en: https://doi.org/10.1038/nmeth.2642
- Koboldt DC. Best practices for variant calling in clinical sequencing. Genome Med [Internet]. 2020;12(1). Disponible en: https://doi.org/10.1186/s13073-020-00791-w
- Maher CA, Kumar-Sinha C, Cao X, Kalyana-Sundaram S, Han B, Jing X, et al. Transcriptome sequencing to detect gene fusions in cancer. Nature [Internet]. 2009;458(7234):97-101. Disponible en: https://doi.org/10.1038/nature07638
- Kogenaru S, Yan Q, Guo Y, Wang N. RNA-seq and microarray complement each other in transcriptome profiling. BMC Genomics [Internet]. 2012;13(1). Disponible en: https://doi.org/10.1186/1471-2164-13-629
- Montgomery SB, Sammeth M, Gutierrez-Arcelus M, Lach RP, Ingle C, Nisbett J, et al. Transcriptome genetics using second generation sequencing in a Caucasian population. Nature [Internet]. 2010;464(7289):773-7. Disponible en: https://doi.org/10.1038/nature08903
- Lawrie CH. MicroRNA expression in lymphoma. Expert Opin Biol Ther [Internet]. 2007;7(9):1363-74. Disponible en: https://doi.org/10.1517/14712598.7.9.1363
- Nelakurthi VM, Paul P, Reche A. Bioinformatics in Early Cancer Detection. Cureus [Internet]. 2023; Disponible en: https://doi.org/10.7759/cureus.46931
- Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, et al. MicroRNA expression profiles classify human cancers. Nature [Internet]. 2005;435(7043):834-8. Disponible en: https://doi.org/10.1038/nature03702
- Van Roosbroeck K, Fanini F, Setoyama T, Ivan C, Rodriguez-Aguayo C, Fuentes-Mattei E, et al. Combining Anti-Mir-155 with Chemotherapy for the Treatment of Lung Cancers. Clin Cancer Res [Internet]. 2016;23(11):2891-904. Disponible en: https://doi.org/10.1158/1078-0432.ccr-16-1025
- Buchberger E, Reis M, Lu TH, Posnien N. Cloudy with a Chance of Insights: Context Dependent Gene Regulation and Implications for Evolutionary Studies. Genes [Internet]. 2019;10(7):492. Disponible en: https://doi.org/10.3390/genes10070492
- Xu M, Yuan L, Wang Y, Chen S, Zhang L, Zhang X. Integrative Analysis of DNA Methylation and Gene Expression Profiles Identifies Colorectal Cancer-Related Diagnostic Biomarkers. Pathol Oncol Res [Internet]. 2021;27. Disponible en: https://doi.org/10.3389/pore.2021.1609784
- Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis [Internet]. 2009;31(1):27-36. Disponible en: https://doi.org/10.1093/carcin/bgp220
- Wilson AS, Power BE, Molloy PL. DNA hypomethylation and human diseases. Biochim Biophys Acta Rev Cancer [Internet]. 2006;1775(1):138-62. Disponible en: https://doi.org/10.1016/j.bbcan.2006.08.007
- Baylin SB. DNA methylation and gene silencing in cancer. Nat Clin Pract Oncol [Internet]. 2005;2(S1):S4-11. Disponible en: https://doi.org/10.1038/ncponc0354
- Manzoor Ahmad Mir, Hina Qayoom, Shazia Sofi, Nusrat Jan, Chapter 2 - Proteomics: A groundbreaking development in cancer biology, Proteomics, Academic Press, 2023, Pages 31-53, https://doi.org/10.1016/B978-0-323-95072-5.00004-3
- Wolde T, Bhardwaj V, Pandey V. Current Bioinformatics Tools in Precision Oncology. MedComm [Internet]. 2025;6(7). Disponible en: https://doi.org/10.1002/mco2.70243
- Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human Breast Cancer: Correlation of Relapse and Survival with Amplification of the HER-2/ neu Oncogene. Science [Internet]. 1987;235(4785):177-82. Disponible en: https://doi.org/10.1126/science.3798106
- Swain SM, Shastry M, Hamilton E. Targeting HER2-positive breast cancer: advances and future directions. Nat Rev Drug Discov [Internet]. 2022;22(2):101-26. Disponible en: https://doi.org/10.1038/s41573-022-00579-0
- Qiu S, Cai Y, Yao H, Lin C, Xie Y, Tang S, et al. Small molecule metabolites: discovery of biomarkers and therapeutic targets. Sig Transduct Target Ther [Internet]. 2023;8(1). Disponible en: https://doi.org/10.1038/s41392-023-01399-3
- Dang L, White DW, Gross S, Bennett BD, Bittinger MA, Driggers EM, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature [Internet]. 2009;462(7274):739-44. Disponible en: https://doi.org/10.1038/nature08617
- Selak MA, Armour SM, MacKenzie ED, Boulahbel H, Watson DG, Mansfield KD, et al. Succinate links TCA cycle dysfunction to oncogenesis by inhibiting HIF-α prolyl hydroxylase. Cancer Cell [Internet]. 2005;7(1):77-85. Disponible en: https://doi.org/10.1016/j.ccr.2004.11.022
- Pollard PJ, Brière JJ, Alam NA, Barwell J, Barclay E, Wortham NC, et al. Accumulation of Krebs cycle intermediates and over-expression of HIF1α in tumours which result from germline FH and SDH mutations. Hum Mol Genet [Internet]. 2005;14(15):2231-9. Disponible en: https://doi.org/10.1093/hmg/ddi227
- Wise DR, DeBerardinis RJ, Mancuso A, Sayed N, Zhang XY, Pfeiffer HK, et al. Myc regulates a transcriptional program that stimulates mitochondrial glutaminolysis and leads to glutamine addiction. Proc Natl Acad Sci U S A [Internet]. 2008;105(48):18782-7. Disponible en: https://doi.org/10.1073/pnas.0810199105
- Wang W, Rong Z, Wang G, Hou Y, Yang F, Qiu M. Cancer metabolites: promising biomarkers for cancer liquid biopsy. Biomarker Res [Internet]. 2023;11(1). Disponible en: https://doi.org/10.1186/s40364-023-00507-3
- Wang G, Qiu M, Xing X, Zhou J, Yao H, Li M, et al. Lung cancer scRNA-seq and lipidomics reveal aberrant lipid metabolism for early-stage diagnosis. Sci Transl Med [Internet]. 2022;14(630). Disponible en: https://doi.org/10.1126/scitranslmed.abk2756
- Ozaki Y, Broughton P, Abdollahi H, Valafar H, Blenda AV. Integrating Omics Data and AI for Cancer Diagnosis and Prognosis. Cancers [Internet]. 2024;16(13):2448. Disponible en: https://doi.org/10.3390/cancers16132448
- Wang ZZ, Li XH, Wen XL, Wang N, Guo Y, Zhu X, et al. Integration of multi-omics data reveals a novel hybrid breast cancer subtype and its biomarkers. Front Oncol [Internet]. 2023;13. Disponible en: https://doi.org/10.3389/fonc.2023.1130092
- Wang N, Li Y, Wang Y, Wang W. Integration of multi-omics profiling reveals an epigenetic-based molecular classification of lung adenocarcinoma: implications for drug sensitivity and immunotherapy response prediction. Front Pharmacol [Internet]. 2025;16. Disponible en: https://doi.org/10.3389/fphar.2025.1540477